Gene
KWMTBOMO02190
Pre Gene Modal
BGIBMGA003133
Annotation
PREDICTED:_ras-related_and_estrogen-regulated_growth_inhibitor-like_protein_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 1.218 Nuclear Reliability : 1.753
Sequence
CDS
ATGAGGGACGAACGGGCTAATGTACCGCGAGTCCGGATTGCCGTCATCGGGAGTTCCAGAGTAGGAAAATCAGCTCTCATTGTTAGATACCTGACTAGAAGATACATAGGCGAATACCATTCAAATACAGATCTCCTGTACAGGCAGACAGTGCCAATAAATGGAACCCCTGTCGAATTAGAAGTGATCGACGTCTCTGGATCAAATTCAGACAAATTCCCCACTGAACAGATACAATGGGCAGACGCCTGCCTCCTTGTGTACGCTGTAACAGACAGGAGCAGCTTTGAATACGCCACAGAAGTTTTGAATGTTTTAAAACGCACTCCTAGCAACGGCAGCCCCGCACATGGAACAACCGTCACATCACCGAGCATGCCCTTAGCACTGCTCGGGAACAAAACTGACCTAGACCATTTGAGACAGGTATCAACAAAAGAAGGTCAGAGTATAAGCGCAGCGCACGGTGCGTCTTTTAGTGAGGCAAGTGTCGCCGACAATTCCGGTGATCTCTACCGCTGCGTTGACCGACTATTGGCCGAAGTTCGCACGCCGCAACGCACGCGAAAATTCTCTGTTACCAAAATGCTTGGCTCGTTAATAGGTGGAGCAACAAATAACAGTCCCACGAACCGTAGCGGGTCAGTCGTAGCGTGCCCGCGGGTGCCGGCGACGTCGCGCGCCCCCTTAGCGGCCGCCGCGCCCTGA
Protein
MRDERANVPRVRIAVIGSSRVGKSALIVRYLTRRYIGEYHSNTDLLYRQTVPINGTPVELEVIDVSGSNSDKFPTEQIQWADACLLVYAVTDRSSFEYATEVLNVLKRTPSNGSPAHGTTVTSPSMPLALLGNKTDLDHLRQVSTKEGQSISAAHGASFSEASVADNSGDLYRCVDRLLAEVRTPQRTRKFSVTKMLGSLIGGATNNSPTNRSGSVVACPRVPATSRAPLAAAAP
Summary
Uniprot
H9J0U6
A0A194R3J5
A0A194PU38
A0A2H1W2Q1
A0A212FNL1
A0A0L7KG49
+ More
A0A1B6HTQ4 A0A1B6L4J1 A0A1B6D1F1 A0A182F2H9 A0A182GPQ1 A0A1B6FFE7 A0A0A9YNH0 A0A1S4F037 A0A1S4H0K9 A0A084WC74 A0A2M4BYV7 A0A182RHC1 A0A182QV44 A0A182JB60 A0A067R521 A0A2M4CQH2 A0A182Y0I8 A0A023F1D8 D6WNF1 A0A1Y1LR50 A0A0T6BHR0 Q7PWF0 A0A336KVV4 A0A224XW86 A0A336MGU1 A0A1W4XHP5 A0A1J1ILH1 A0A1B0EVY1 A0A2M4CVF6 A0A034WHV6 A0A1A9WZY0 A0A1A9XZI1 A0A0L0CCI8 A0A182LMI1 A0A182NBL0 A0A182I7K1 A0A0K8V1Q2 A0A1B0FIZ3 A0A182VMW9 Q17JX3 A0A1A9Z7S5 T1PD59 A0A182KG13 A0A182WGD5 W8BWV2 A0A1I8P738 A0A182WXN6 A0A1B0D3I5 W5JBR9 A0A0K8W9N3 A0A1A9UMI0 A0A2P8XMX3 A0A0P6AC93 A0A226D4D1 A0A1B6J9R2 A0A226E4F1 A0A1Y1LJL4 A0A0P5F5R3 A0A226E1W2 A0A0P5N4D3 A0A0P6EWK7 A0A226D4H1 A0A087ZQC6 A0A232FFQ6 K7JAF9 A0A3Q0JCU6 A0A158NH60 A0A3L8DMZ9 A0A151X5I1 A0A0P5IFW7 A0A023F4H7 A0A1B6GMD8 A0A0L7R967 A0A0P5BCF3 A0A195D838 A0A195E8D3 A0A158NFE4 A0A151XFV6 J9JW93 A0A0P4WJ18 A0A2S2NEQ1 A0A084VYU1 A0A182J4U6 A0A1S4GIE3 A0A182KXI5 A0A0L0CR13 E2A6B4 A0A0K8VY33 A0A3L8DGC7 N6TZS7 U4UF97
A0A1B6HTQ4 A0A1B6L4J1 A0A1B6D1F1 A0A182F2H9 A0A182GPQ1 A0A1B6FFE7 A0A0A9YNH0 A0A1S4F037 A0A1S4H0K9 A0A084WC74 A0A2M4BYV7 A0A182RHC1 A0A182QV44 A0A182JB60 A0A067R521 A0A2M4CQH2 A0A182Y0I8 A0A023F1D8 D6WNF1 A0A1Y1LR50 A0A0T6BHR0 Q7PWF0 A0A336KVV4 A0A224XW86 A0A336MGU1 A0A1W4XHP5 A0A1J1ILH1 A0A1B0EVY1 A0A2M4CVF6 A0A034WHV6 A0A1A9WZY0 A0A1A9XZI1 A0A0L0CCI8 A0A182LMI1 A0A182NBL0 A0A182I7K1 A0A0K8V1Q2 A0A1B0FIZ3 A0A182VMW9 Q17JX3 A0A1A9Z7S5 T1PD59 A0A182KG13 A0A182WGD5 W8BWV2 A0A1I8P738 A0A182WXN6 A0A1B0D3I5 W5JBR9 A0A0K8W9N3 A0A1A9UMI0 A0A2P8XMX3 A0A0P6AC93 A0A226D4D1 A0A1B6J9R2 A0A226E4F1 A0A1Y1LJL4 A0A0P5F5R3 A0A226E1W2 A0A0P5N4D3 A0A0P6EWK7 A0A226D4H1 A0A087ZQC6 A0A232FFQ6 K7JAF9 A0A3Q0JCU6 A0A158NH60 A0A3L8DMZ9 A0A151X5I1 A0A0P5IFW7 A0A023F4H7 A0A1B6GMD8 A0A0L7R967 A0A0P5BCF3 A0A195D838 A0A195E8D3 A0A158NFE4 A0A151XFV6 J9JW93 A0A0P4WJ18 A0A2S2NEQ1 A0A084VYU1 A0A182J4U6 A0A1S4GIE3 A0A182KXI5 A0A0L0CR13 E2A6B4 A0A0K8VY33 A0A3L8DGC7 N6TZS7 U4UF97
Pubmed
EMBL
BABH01010366
KQ460779
KPJ12267.1
KQ459598
KPI94650.1
ODYU01005937
+ More
SOQ47351.1 AGBW02005153 OWR55336.1 JTDY01009998 KOB61879.1 GECU01029649 GECU01020352 GECU01011879 GECU01001993 JAS78057.1 JAS87354.1 JAS95827.1 JAT05714.1 GEBQ01021391 JAT18586.1 GEDC01028352 GEDC01017767 JAS08946.1 JAS19531.1 JXUM01078889 JXUM01078890 JXUM01078891 JXUM01078892 JXUM01078893 GECZ01020852 GECZ01001795 JAS48917.1 JAS67974.1 GBHO01010433 GBRD01017042 GDHC01011423 JAG33171.1 JAG48785.1 JAQ07206.1 AAAB01008984 ATLV01022579 KE525333 KFB47818.1 GGFJ01009109 MBW58250.1 AXCN02000676 KK852691 KDR18359.1 GGFL01003253 MBW67431.1 GBBI01003422 JAC15290.1 KQ971343 EFA03227.1 GEZM01054655 JAV73507.1 LJIG01000241 KRT86699.1 EAA15139.3 UFQS01001049 UFQT01001049 SSX08718.1 SSX28630.1 GFTR01004033 JAW12393.1 UFQT01000560 SSX25308.1 CVRI01000055 CRL01087.1 AJWK01022187 AJWK01022188 GGFL01005067 MBW69245.1 GAKP01005025 JAC53927.1 JRES01000682 KNC29174.1 APCN01003411 GDHF01019558 JAI32756.1 CCAG010013969 CCAG010013970 CCAG010013971 CH477230 EAT46938.1 KA646695 KA646696 AFP61325.1 GAMC01012879 GAMC01012876 JAB93679.1 AJVK01010987 AJVK01010988 AJVK01010989 AJVK01010990 ADMH02001853 ETN60858.1 GDHF01004744 JAI47570.1 PYGN01001685 PSN33358.1 GDIP01031854 JAM71861.1 LNIX01000033 OXA40412.1 GECU01011841 JAS95865.1 LNIX01000007 OXA51366.1 GEZM01054090 JAV73852.1 GDIQ01259969 JAJ91755.1 OXA51543.1 GDIQ01169378 JAK82347.1 GDIQ01069663 JAN25074.1 OXA40452.1 NNAY01000303 OXU29350.1 ADTU01015589 ADTU01015590 ADTU01015591 ADTU01015592 ADTU01015593 QOIP01000006 RLU21797.1 KQ982510 KYQ55627.1 GDIQ01238599 JAK13126.1 GBBI01002286 JAC16426.1 GECZ01006208 JAS63561.1 KQ414627 KOC67415.1 GDIP01186392 LRGB01002190 JAJ37010.1 KZS08644.1 KQ976750 KYN08614.1 KQ979440 KYN21475.1 ADTU01014359 KQ982182 KYQ59160.1 ABLF02034324 ABLF02034329 ABLF02034331 ABLF02034332 GDRN01041774 GDRN01041773 JAI67087.1 GGMR01003031 MBY15650.1 ATLV01018464 ATLV01018465 ATLV01018466 KE525234 KFB43135.1 AXCP01003532 AXCP01003533 AAAB01008888 JRES01000036 KNC34686.1 GL437123 EFN70999.1 GDHF01019003 GDHF01017294 GDHF01008578 JAI33311.1 JAI35020.1 JAI43736.1 QOIP01000009 RLU18969.1 APGK01044802 APGK01044803 APGK01044804 APGK01044805 KB741028 ENN74800.1 KB632297 ERL91707.1
SOQ47351.1 AGBW02005153 OWR55336.1 JTDY01009998 KOB61879.1 GECU01029649 GECU01020352 GECU01011879 GECU01001993 JAS78057.1 JAS87354.1 JAS95827.1 JAT05714.1 GEBQ01021391 JAT18586.1 GEDC01028352 GEDC01017767 JAS08946.1 JAS19531.1 JXUM01078889 JXUM01078890 JXUM01078891 JXUM01078892 JXUM01078893 GECZ01020852 GECZ01001795 JAS48917.1 JAS67974.1 GBHO01010433 GBRD01017042 GDHC01011423 JAG33171.1 JAG48785.1 JAQ07206.1 AAAB01008984 ATLV01022579 KE525333 KFB47818.1 GGFJ01009109 MBW58250.1 AXCN02000676 KK852691 KDR18359.1 GGFL01003253 MBW67431.1 GBBI01003422 JAC15290.1 KQ971343 EFA03227.1 GEZM01054655 JAV73507.1 LJIG01000241 KRT86699.1 EAA15139.3 UFQS01001049 UFQT01001049 SSX08718.1 SSX28630.1 GFTR01004033 JAW12393.1 UFQT01000560 SSX25308.1 CVRI01000055 CRL01087.1 AJWK01022187 AJWK01022188 GGFL01005067 MBW69245.1 GAKP01005025 JAC53927.1 JRES01000682 KNC29174.1 APCN01003411 GDHF01019558 JAI32756.1 CCAG010013969 CCAG010013970 CCAG010013971 CH477230 EAT46938.1 KA646695 KA646696 AFP61325.1 GAMC01012879 GAMC01012876 JAB93679.1 AJVK01010987 AJVK01010988 AJVK01010989 AJVK01010990 ADMH02001853 ETN60858.1 GDHF01004744 JAI47570.1 PYGN01001685 PSN33358.1 GDIP01031854 JAM71861.1 LNIX01000033 OXA40412.1 GECU01011841 JAS95865.1 LNIX01000007 OXA51366.1 GEZM01054090 JAV73852.1 GDIQ01259969 JAJ91755.1 OXA51543.1 GDIQ01169378 JAK82347.1 GDIQ01069663 JAN25074.1 OXA40452.1 NNAY01000303 OXU29350.1 ADTU01015589 ADTU01015590 ADTU01015591 ADTU01015592 ADTU01015593 QOIP01000006 RLU21797.1 KQ982510 KYQ55627.1 GDIQ01238599 JAK13126.1 GBBI01002286 JAC16426.1 GECZ01006208 JAS63561.1 KQ414627 KOC67415.1 GDIP01186392 LRGB01002190 JAJ37010.1 KZS08644.1 KQ976750 KYN08614.1 KQ979440 KYN21475.1 ADTU01014359 KQ982182 KYQ59160.1 ABLF02034324 ABLF02034329 ABLF02034331 ABLF02034332 GDRN01041774 GDRN01041773 JAI67087.1 GGMR01003031 MBY15650.1 ATLV01018464 ATLV01018465 ATLV01018466 KE525234 KFB43135.1 AXCP01003532 AXCP01003533 AAAB01008888 JRES01000036 KNC34686.1 GL437123 EFN70999.1 GDHF01019003 GDHF01017294 GDHF01008578 JAI33311.1 JAI35020.1 JAI43736.1 QOIP01000009 RLU18969.1 APGK01044802 APGK01044803 APGK01044804 APGK01044805 KB741028 ENN74800.1 KB632297 ERL91707.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000069272
+ More
UP000069940 UP000030765 UP000075900 UP000075886 UP000075880 UP000027135 UP000076408 UP000007266 UP000007062 UP000192223 UP000183832 UP000092461 UP000091820 UP000092443 UP000037069 UP000075882 UP000075884 UP000075840 UP000092444 UP000075903 UP000008820 UP000092445 UP000095301 UP000075881 UP000075920 UP000095300 UP000076407 UP000092462 UP000000673 UP000078200 UP000245037 UP000198287 UP000005203 UP000215335 UP000002358 UP000079169 UP000005205 UP000279307 UP000075809 UP000053825 UP000076858 UP000078542 UP000078492 UP000007819 UP000000311 UP000019118 UP000030742
UP000069940 UP000030765 UP000075900 UP000075886 UP000075880 UP000027135 UP000076408 UP000007266 UP000007062 UP000192223 UP000183832 UP000092461 UP000091820 UP000092443 UP000037069 UP000075882 UP000075884 UP000075840 UP000092444 UP000075903 UP000008820 UP000092445 UP000095301 UP000075881 UP000075920 UP000095300 UP000076407 UP000092462 UP000000673 UP000078200 UP000245037 UP000198287 UP000005203 UP000215335 UP000002358 UP000079169 UP000005205 UP000279307 UP000075809 UP000053825 UP000076858 UP000078542 UP000078492 UP000007819 UP000000311 UP000019118 UP000030742
PRIDE
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
H9J0U6
A0A194R3J5
A0A194PU38
A0A2H1W2Q1
A0A212FNL1
A0A0L7KG49
+ More
A0A1B6HTQ4 A0A1B6L4J1 A0A1B6D1F1 A0A182F2H9 A0A182GPQ1 A0A1B6FFE7 A0A0A9YNH0 A0A1S4F037 A0A1S4H0K9 A0A084WC74 A0A2M4BYV7 A0A182RHC1 A0A182QV44 A0A182JB60 A0A067R521 A0A2M4CQH2 A0A182Y0I8 A0A023F1D8 D6WNF1 A0A1Y1LR50 A0A0T6BHR0 Q7PWF0 A0A336KVV4 A0A224XW86 A0A336MGU1 A0A1W4XHP5 A0A1J1ILH1 A0A1B0EVY1 A0A2M4CVF6 A0A034WHV6 A0A1A9WZY0 A0A1A9XZI1 A0A0L0CCI8 A0A182LMI1 A0A182NBL0 A0A182I7K1 A0A0K8V1Q2 A0A1B0FIZ3 A0A182VMW9 Q17JX3 A0A1A9Z7S5 T1PD59 A0A182KG13 A0A182WGD5 W8BWV2 A0A1I8P738 A0A182WXN6 A0A1B0D3I5 W5JBR9 A0A0K8W9N3 A0A1A9UMI0 A0A2P8XMX3 A0A0P6AC93 A0A226D4D1 A0A1B6J9R2 A0A226E4F1 A0A1Y1LJL4 A0A0P5F5R3 A0A226E1W2 A0A0P5N4D3 A0A0P6EWK7 A0A226D4H1 A0A087ZQC6 A0A232FFQ6 K7JAF9 A0A3Q0JCU6 A0A158NH60 A0A3L8DMZ9 A0A151X5I1 A0A0P5IFW7 A0A023F4H7 A0A1B6GMD8 A0A0L7R967 A0A0P5BCF3 A0A195D838 A0A195E8D3 A0A158NFE4 A0A151XFV6 J9JW93 A0A0P4WJ18 A0A2S2NEQ1 A0A084VYU1 A0A182J4U6 A0A1S4GIE3 A0A182KXI5 A0A0L0CR13 E2A6B4 A0A0K8VY33 A0A3L8DGC7 N6TZS7 U4UF97
A0A1B6HTQ4 A0A1B6L4J1 A0A1B6D1F1 A0A182F2H9 A0A182GPQ1 A0A1B6FFE7 A0A0A9YNH0 A0A1S4F037 A0A1S4H0K9 A0A084WC74 A0A2M4BYV7 A0A182RHC1 A0A182QV44 A0A182JB60 A0A067R521 A0A2M4CQH2 A0A182Y0I8 A0A023F1D8 D6WNF1 A0A1Y1LR50 A0A0T6BHR0 Q7PWF0 A0A336KVV4 A0A224XW86 A0A336MGU1 A0A1W4XHP5 A0A1J1ILH1 A0A1B0EVY1 A0A2M4CVF6 A0A034WHV6 A0A1A9WZY0 A0A1A9XZI1 A0A0L0CCI8 A0A182LMI1 A0A182NBL0 A0A182I7K1 A0A0K8V1Q2 A0A1B0FIZ3 A0A182VMW9 Q17JX3 A0A1A9Z7S5 T1PD59 A0A182KG13 A0A182WGD5 W8BWV2 A0A1I8P738 A0A182WXN6 A0A1B0D3I5 W5JBR9 A0A0K8W9N3 A0A1A9UMI0 A0A2P8XMX3 A0A0P6AC93 A0A226D4D1 A0A1B6J9R2 A0A226E4F1 A0A1Y1LJL4 A0A0P5F5R3 A0A226E1W2 A0A0P5N4D3 A0A0P6EWK7 A0A226D4H1 A0A087ZQC6 A0A232FFQ6 K7JAF9 A0A3Q0JCU6 A0A158NH60 A0A3L8DMZ9 A0A151X5I1 A0A0P5IFW7 A0A023F4H7 A0A1B6GMD8 A0A0L7R967 A0A0P5BCF3 A0A195D838 A0A195E8D3 A0A158NFE4 A0A151XFV6 J9JW93 A0A0P4WJ18 A0A2S2NEQ1 A0A084VYU1 A0A182J4U6 A0A1S4GIE3 A0A182KXI5 A0A0L0CR13 E2A6B4 A0A0K8VY33 A0A3L8DGC7 N6TZS7 U4UF97
PDB
2ATV
E-value=5.19334e-25,
Score=281
Ontologies
KEGG
GO
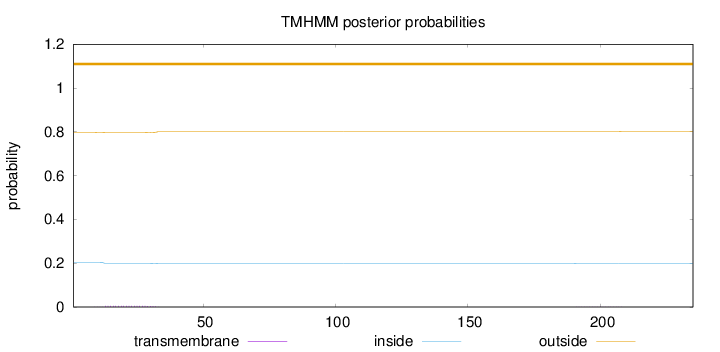
Topology
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0991599999999998
Exp number, first 60 AAs:
0.09384
Total prob of N-in:
0.20328
outside
1 - 235
Population Genetic Test Statistics
Pi
196.470881
Theta
192.705832
Tajima's D
0.220016
CLR
1.937133
CSRT
0.433628318584071
Interpretation
Uncertain
