Gene
KWMTBOMO02189
Pre Gene Modal
BGIBMGA003134
Annotation
PREDICTED:_class_E_basic_helix-loop-helix_protein_22-like_[Papilio_polytes]
Transcription factor
Location in the cell
Nuclear Reliability : 3.49
Sequence
CDS
ATGGAGGGGCGGAGGAATTGGGATGGTGTACCTTTGGGCGAGTCGCATTCGCCGCCGCAGTCGGTGCCTGGTCGAAGGACGCCGCTTGGAGCCGTGGGCCTCGGTGGCTTTTACATGCAAGGAGGACAGCCTCCGCCCCCACAACATTGTTATCCTACGGACGAGAACCAGCCAGAACCAGGAACCAGCTATGGACAAAGGCCAGTCAACGAAAGTAAATCGAAACATAAGATGCGACAAGGTAAAACTGTGCGCCTGAACATAAACGCCCGTGAACGAAGACGTATGCACGATTTAAATGACGCCTTGGACGAACTCCGAGGTGTGATACCATACGCTCATTCGCCGTCCGTACGAAAGCTGTCAAAGATAGCAACGCTTTTATTGGCCAAGAATTATATTATGATGCAGGCTAGCGCGTTAGAAGAACTACGAAGACTTGTAGCGTACTTGCAAGGTACGGCAACAGCGGCGGGTATTGTGCCCCCACCGGGTTTCGATCTAGCCGGTTTTCCAGCGGCAGCCAAGCTCCTTCAGCCTCCAGCCCCGGGCCCTCCACCGCCCCCAGAACCACCCTCTTGA
Protein
MEGRRNWDGVPLGESHSPPQSVPGRRTPLGAVGLGGFYMQGGQPPPPQHCYPTDENQPEPGTSYGQRPVNESKSKHKMRQGKTVRLNINARERRRMHDLNDALDELRGVIPYAHSPSVRKLSKIATLLLAKNYIMMQASALEELRRLVAYLQGTATAAGIVPPPGFDLAGFPAAAKLLQPPAPGPPPPPEPPS
Summary
Uniprot
H9J0U7
A0A194PMJ1
A0A194R385
A0A2A4IXY1
A0A2H1W2M0
D6WP00
+ More
A0A0K0PQL7 A0A1W4XPV7 U4TS00 U4U7Q7 T1GGJ1 A0A1B0CX77 A0A1W4UYK6 A0A0K8UQV3 A0A034WDH9 A0A0A1WYX2 B4I5C5 B3NMF7 B4P8Q1 B4Q800 Q9VJC1 B3MMG5 B4KEE4 A0A3B0JI90 B4LSV0 A0A0M5IW58 B4MYV4 A0A1S4H1A1 A0A182VFT7 A0A182KUA3 A0A182PAM2 A0A182TTP4 A0A182I5A8 A0A182XKT6 B4GQ98 Q29LC0 A0A2H8TXE3 W5JFG7 A0A336MMB8 B0W182 A0A182F123 A0A026X5J4 A0A1S3D3L0 A0A182WHA6 Q17PH4 A0A182RG52 E0VY24 A0A182ISR1 J9JJL3 A0A2S2N8I4 Q5TNI3 A0A2S2QD44 A0A0L0BQL9 A0A182KEL5 A0A1B6E7Y3 A0A182Y9V7 A0A182QVM2 A0A182NF84 A0A154P6X5 A0A088A1F2 A0A0N0BHQ4 B4JC94 E2C838 E2AGK1 A0A151I618 E9J396 A0A151HZR0 A0A158NLU1 A0A1J1HEQ4 A0A310SI37 A0A0K8UNN3 A0A1A9WLM7 A0A1B0FN87 A0A232EVK5 A0A1A9VQC7 A0A1B0AJM1 K7IQT8 A0A1I8M4F6 T1HBE9 A0A2R7X1H8 A0A182LUS1 A0A1A9Y771 A0A067QHI1 A0A1B6L3K7 F4WU54
A0A0K0PQL7 A0A1W4XPV7 U4TS00 U4U7Q7 T1GGJ1 A0A1B0CX77 A0A1W4UYK6 A0A0K8UQV3 A0A034WDH9 A0A0A1WYX2 B4I5C5 B3NMF7 B4P8Q1 B4Q800 Q9VJC1 B3MMG5 B4KEE4 A0A3B0JI90 B4LSV0 A0A0M5IW58 B4MYV4 A0A1S4H1A1 A0A182VFT7 A0A182KUA3 A0A182PAM2 A0A182TTP4 A0A182I5A8 A0A182XKT6 B4GQ98 Q29LC0 A0A2H8TXE3 W5JFG7 A0A336MMB8 B0W182 A0A182F123 A0A026X5J4 A0A1S3D3L0 A0A182WHA6 Q17PH4 A0A182RG52 E0VY24 A0A182ISR1 J9JJL3 A0A2S2N8I4 Q5TNI3 A0A2S2QD44 A0A0L0BQL9 A0A182KEL5 A0A1B6E7Y3 A0A182Y9V7 A0A182QVM2 A0A182NF84 A0A154P6X5 A0A088A1F2 A0A0N0BHQ4 B4JC94 E2C838 E2AGK1 A0A151I618 E9J396 A0A151HZR0 A0A158NLU1 A0A1J1HEQ4 A0A310SI37 A0A0K8UNN3 A0A1A9WLM7 A0A1B0FN87 A0A232EVK5 A0A1A9VQC7 A0A1B0AJM1 K7IQT8 A0A1I8M4F6 T1HBE9 A0A2R7X1H8 A0A182LUS1 A0A1A9Y771 A0A067QHI1 A0A1B6L3K7 F4WU54
Pubmed
19121390
26354079
18362917
19820115
23537049
25348373
+ More
25830018 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 12364791 20966253 15632085 23185243 20920257 23761445 24508170 30249741 17510324 20566863 26108605 25244985 20798317 21282665 21347285 28648823 20075255 25315136 24845553 21719571
25830018 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 12364791 20966253 15632085 23185243 20920257 23761445 24508170 30249741 17510324 20566863 26108605 25244985 20798317 21282665 21347285 28648823 20075255 25315136 24845553 21719571
EMBL
BABH01010367
KQ459598
KPI94651.1
KQ460779
KPJ12268.1
NWSH01004924
+ More
PCG64581.1 ODYU01005937 SOQ47350.1 KQ971343 EFA04412.1 KP641327 AKQ48969.1 KB630267 KB631411 ERL83502.1 ERL84284.1 KB632146 ERL89102.1 CAQQ02195770 CAQQ02195771 AJWK01033330 GDHF01023247 GDHF01003831 JAI29067.1 JAI48483.1 GAKP01005321 JAC53631.1 GBXI01010245 JAD04047.1 CH480822 EDW55581.1 CH954179 EDV54828.1 CM000158 EDW90159.1 CM000361 CM002910 EDX05333.1 KMY90732.1 AE014134 BT004905 AAF53631.1 AAO47883.1 ADV37079.1 ADV37080.1 AHN54531.1 CH902620 EDV30911.1 CH933807 EDW12912.1 KRG03482.1 KRG03483.1 OUUW01000004 SPP80022.1 CH940649 EDW64861.1 KRF81898.1 CP012523 ALC38674.1 CH963913 EDW77293.1 AAAB01008984 APCN01003844 CH479187 EDW39770.1 CH379060 EAL34125.2 KRT04507.1 GFXV01001829 GFXV01004907 GFXV01006033 GFXV01006701 MBW13634.1 MBW16712.1 MBW17838.1 MBW18506.1 ADMH02001645 ETN61625.1 UFQT01001365 SSX30219.1 DS231820 EDS44056.1 KK107019 QOIP01000007 EZA62714.1 RLU20022.1 CH477191 EAT48643.1 DS235843 EEB18280.1 ABLF02028130 GGMR01000821 MBY13440.1 EAL39075.2 GGMS01006454 MBY75657.1 JRES01001610 KNC21519.1 GEDC01003258 JAS34040.1 AXCN02000825 KQ434819 KZC06880.1 KQ435748 KOX76422.1 CH916368 EDW04127.1 GL453538 EFN75891.1 GL439316 EFN67458.1 KQ978524 KYM93368.1 GL768059 EFZ12709.1 KQ976699 KYM77422.1 ADTU01019857 ADTU01019858 CVRI01000001 CRK86431.1 KQ765234 OAD54000.1 GDHF01024030 GDHF01014144 JAI28284.1 JAI38170.1 CCAG010014258 NNAY01001979 OXU22393.1 ACPB03008942 KK856368 PTY25616.1 AXCM01011269 KK853378 KDR08028.1 GEBQ01021756 JAT18221.1 GL888349 EGI62281.1
PCG64581.1 ODYU01005937 SOQ47350.1 KQ971343 EFA04412.1 KP641327 AKQ48969.1 KB630267 KB631411 ERL83502.1 ERL84284.1 KB632146 ERL89102.1 CAQQ02195770 CAQQ02195771 AJWK01033330 GDHF01023247 GDHF01003831 JAI29067.1 JAI48483.1 GAKP01005321 JAC53631.1 GBXI01010245 JAD04047.1 CH480822 EDW55581.1 CH954179 EDV54828.1 CM000158 EDW90159.1 CM000361 CM002910 EDX05333.1 KMY90732.1 AE014134 BT004905 AAF53631.1 AAO47883.1 ADV37079.1 ADV37080.1 AHN54531.1 CH902620 EDV30911.1 CH933807 EDW12912.1 KRG03482.1 KRG03483.1 OUUW01000004 SPP80022.1 CH940649 EDW64861.1 KRF81898.1 CP012523 ALC38674.1 CH963913 EDW77293.1 AAAB01008984 APCN01003844 CH479187 EDW39770.1 CH379060 EAL34125.2 KRT04507.1 GFXV01001829 GFXV01004907 GFXV01006033 GFXV01006701 MBW13634.1 MBW16712.1 MBW17838.1 MBW18506.1 ADMH02001645 ETN61625.1 UFQT01001365 SSX30219.1 DS231820 EDS44056.1 KK107019 QOIP01000007 EZA62714.1 RLU20022.1 CH477191 EAT48643.1 DS235843 EEB18280.1 ABLF02028130 GGMR01000821 MBY13440.1 EAL39075.2 GGMS01006454 MBY75657.1 JRES01001610 KNC21519.1 GEDC01003258 JAS34040.1 AXCN02000825 KQ434819 KZC06880.1 KQ435748 KOX76422.1 CH916368 EDW04127.1 GL453538 EFN75891.1 GL439316 EFN67458.1 KQ978524 KYM93368.1 GL768059 EFZ12709.1 KQ976699 KYM77422.1 ADTU01019857 ADTU01019858 CVRI01000001 CRK86431.1 KQ765234 OAD54000.1 GDHF01024030 GDHF01014144 JAI28284.1 JAI38170.1 CCAG010014258 NNAY01001979 OXU22393.1 ACPB03008942 KK856368 PTY25616.1 AXCM01011269 KK853378 KDR08028.1 GEBQ01021756 JAT18221.1 GL888349 EGI62281.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007266
UP000192223
+ More
UP000030742 UP000015102 UP000092461 UP000192221 UP000001292 UP000008711 UP000002282 UP000000304 UP000000803 UP000007801 UP000009192 UP000268350 UP000008792 UP000092553 UP000007798 UP000075903 UP000075882 UP000075885 UP000075902 UP000075840 UP000076407 UP000008744 UP000001819 UP000000673 UP000002320 UP000069272 UP000053097 UP000279307 UP000079169 UP000075920 UP000008820 UP000075900 UP000009046 UP000075880 UP000007819 UP000007062 UP000037069 UP000075881 UP000076408 UP000075886 UP000075884 UP000076502 UP000005203 UP000053105 UP000001070 UP000008237 UP000000311 UP000078542 UP000078540 UP000005205 UP000183832 UP000091820 UP000092444 UP000215335 UP000078200 UP000092445 UP000002358 UP000095301 UP000015103 UP000075883 UP000092443 UP000027135 UP000007755
UP000030742 UP000015102 UP000092461 UP000192221 UP000001292 UP000008711 UP000002282 UP000000304 UP000000803 UP000007801 UP000009192 UP000268350 UP000008792 UP000092553 UP000007798 UP000075903 UP000075882 UP000075885 UP000075902 UP000075840 UP000076407 UP000008744 UP000001819 UP000000673 UP000002320 UP000069272 UP000053097 UP000279307 UP000079169 UP000075920 UP000008820 UP000075900 UP000009046 UP000075880 UP000007819 UP000007062 UP000037069 UP000075881 UP000076408 UP000075886 UP000075884 UP000076502 UP000005203 UP000053105 UP000001070 UP000008237 UP000000311 UP000078542 UP000078540 UP000005205 UP000183832 UP000091820 UP000092444 UP000215335 UP000078200 UP000092445 UP000002358 UP000095301 UP000015103 UP000075883 UP000092443 UP000027135 UP000007755
Interpro
SUPFAM
SSF47459
SSF47459
Gene 3D
CDD
ProteinModelPortal
H9J0U7
A0A194PMJ1
A0A194R385
A0A2A4IXY1
A0A2H1W2M0
D6WP00
+ More
A0A0K0PQL7 A0A1W4XPV7 U4TS00 U4U7Q7 T1GGJ1 A0A1B0CX77 A0A1W4UYK6 A0A0K8UQV3 A0A034WDH9 A0A0A1WYX2 B4I5C5 B3NMF7 B4P8Q1 B4Q800 Q9VJC1 B3MMG5 B4KEE4 A0A3B0JI90 B4LSV0 A0A0M5IW58 B4MYV4 A0A1S4H1A1 A0A182VFT7 A0A182KUA3 A0A182PAM2 A0A182TTP4 A0A182I5A8 A0A182XKT6 B4GQ98 Q29LC0 A0A2H8TXE3 W5JFG7 A0A336MMB8 B0W182 A0A182F123 A0A026X5J4 A0A1S3D3L0 A0A182WHA6 Q17PH4 A0A182RG52 E0VY24 A0A182ISR1 J9JJL3 A0A2S2N8I4 Q5TNI3 A0A2S2QD44 A0A0L0BQL9 A0A182KEL5 A0A1B6E7Y3 A0A182Y9V7 A0A182QVM2 A0A182NF84 A0A154P6X5 A0A088A1F2 A0A0N0BHQ4 B4JC94 E2C838 E2AGK1 A0A151I618 E9J396 A0A151HZR0 A0A158NLU1 A0A1J1HEQ4 A0A310SI37 A0A0K8UNN3 A0A1A9WLM7 A0A1B0FN87 A0A232EVK5 A0A1A9VQC7 A0A1B0AJM1 K7IQT8 A0A1I8M4F6 T1HBE9 A0A2R7X1H8 A0A182LUS1 A0A1A9Y771 A0A067QHI1 A0A1B6L3K7 F4WU54
A0A0K0PQL7 A0A1W4XPV7 U4TS00 U4U7Q7 T1GGJ1 A0A1B0CX77 A0A1W4UYK6 A0A0K8UQV3 A0A034WDH9 A0A0A1WYX2 B4I5C5 B3NMF7 B4P8Q1 B4Q800 Q9VJC1 B3MMG5 B4KEE4 A0A3B0JI90 B4LSV0 A0A0M5IW58 B4MYV4 A0A1S4H1A1 A0A182VFT7 A0A182KUA3 A0A182PAM2 A0A182TTP4 A0A182I5A8 A0A182XKT6 B4GQ98 Q29LC0 A0A2H8TXE3 W5JFG7 A0A336MMB8 B0W182 A0A182F123 A0A026X5J4 A0A1S3D3L0 A0A182WHA6 Q17PH4 A0A182RG52 E0VY24 A0A182ISR1 J9JJL3 A0A2S2N8I4 Q5TNI3 A0A2S2QD44 A0A0L0BQL9 A0A182KEL5 A0A1B6E7Y3 A0A182Y9V7 A0A182QVM2 A0A182NF84 A0A154P6X5 A0A088A1F2 A0A0N0BHQ4 B4JC94 E2C838 E2AGK1 A0A151I618 E9J396 A0A151HZR0 A0A158NLU1 A0A1J1HEQ4 A0A310SI37 A0A0K8UNN3 A0A1A9WLM7 A0A1B0FN87 A0A232EVK5 A0A1A9VQC7 A0A1B0AJM1 K7IQT8 A0A1I8M4F6 T1HBE9 A0A2R7X1H8 A0A182LUS1 A0A1A9Y771 A0A067QHI1 A0A1B6L3K7 F4WU54
PDB
2QL2
E-value=1.17753e-08,
Score=138
Ontologies
GO
PANTHER
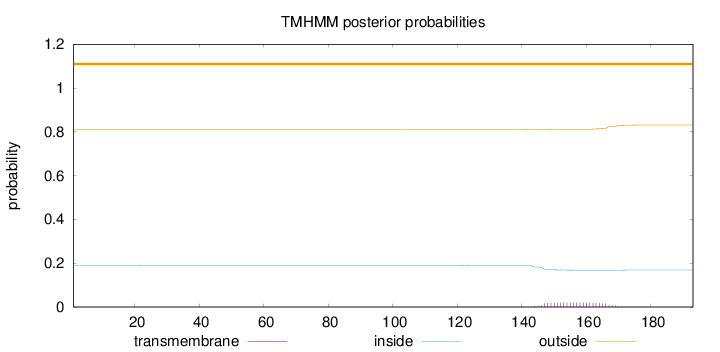
Topology
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.45324
Exp number, first 60 AAs:
0.00091
Total prob of N-in:
0.18921
outside
1 - 193
Population Genetic Test Statistics
Pi
215.335375
Theta
161.270851
Tajima's D
1.059953
CLR
0.73811
CSRT
0.674416279186041
Interpretation
Uncertain
