Gene
KWMTBOMO02178
Pre Gene Modal
BGIBMGA003025
Annotation
PREDICTED:_chymotrypsin-1-like_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 2.438
Sequence
CDS
ATGGAGGGTAAAGTGGTAGGAGGGAAATCAGGTTCTCTCGAGCAGTTTCCTTTTCAAGCACAAGTATACAATTTAGGTGCTATTTGTGCTGGGACGATATTAAACAGCTGGACAATACTGACGGTGGCTCATTGCTTTGATCACAATAAGAATGTCCGAGAGTTGGAAGTGCAGACGGGTTCAAAATACTTATATGGCTTCAAAGCAAAAAAGTATAACGTCAGTTCCTTGGTCGTTCACAAAGCGTATAACACGACAGCTCAATTCAGCAACGACATTGCTCTGATCTTCTTGACTAGTCCCGTAGAATTCGGGAAGCTAGTGCAGAAAGGTATTCTGGTTGATAATGGAAAATGGATGTCTAAGTATGAAAAAAACTTTATTGTCAGTGGCTGGGGATGGACACAGTATGGAGGGCCAATATCACAGCGTGGATTACTGACGACACATCTGGGATACGTTCCATCTCGGCAGTGTGGTAGATTTCACAATCTCAGCCTCACCCCAGACATGTTTTGTTTGTATGGGGATGGTGAAAGGGATACCTGTAAGGGTGATTCAGGCACTGGAGTACTTTGGAATGGGTACATCGTGGGGATTACTTCACACGGCGACGGCTGCGCGAAGAAATATAAGCCGAGTGTTTATACCAACGTTTGGTATTTCAGGGACTGGATCGAAAAACATATAAACAATTTTTTTAAGGCGTTTTGCAAGTTTAACCAAACATCCGTACAAAAGAAATTAAAGACGAAAACGTAA
Protein
MEGKVVGGKSGSLEQFPFQAQVYNLGAICAGTILNSWTILTVAHCFDHNKNVRELEVQTGSKYLYGFKAKKYNVSSLVVHKAYNTTAQFSNDIALIFLTSPVEFGKLVQKGILVDNGKWMSKYEKNFIVSGWGWTQYGGPISQRGLLTTHLGYVPSRQCGRFHNLSLTPDMFCLYGDGERDTCKGDSGTGVLWNGYIVGITSHGDGCAKKYKPSVYTNVWYFRDWIEKHINNFFKAFCKFNQTSVQKKLKTKT
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9J0I8
A0A194R398
A0A2A4JWI7
A0A212F7S3
A0A194PMK0
A0A0L7LH96
+ More
A0A0A2KWH5 A0A226D5J2 A0A0M9A9Z8 A0A3D8S5S2 A0A0M5IZZ2 A0A232EXU8 E9H014 Q8MNY6 A0A068F5B8 A0A0P5XLE4 A0A3M6TQ33 A0A0P5VLQ4 A0A0N8CRT0 B4JVV3 A0A0P4YLS3 A0A0N8BDG1 A0A0N8CYB7 A0A212FL72 A0A194R423 A0A3B0JSD8 A0A0P5V5F0 A0A0P5GPW3 A0A0N8AVR2 A0A182GGY4 A0A0P5FVM8 A0A2I3SE40 A0A0P6BEL6 A0A0P5JH56 A0A0P5QQH8 A0A0P5GA08 A0A0P6AQ80 A0A1L8HMQ5 A0A0P6E6W6 G3S3C4 A0A0P5L024 A0A1W7R806 A0A0P5MDH3 Q5QBF4 A0A1I8Q0B3 A0A0P5GX76 A0A0P6F713 K7J4L1 A0A0P5J6E5 A0A0P5GGK0 A0A0P5GL06 A0A0P5FRW8 A0A0P5GQL0 A0A232FET9 F6XTG5 A0A0P5KPC1 A0A0P5G8N5 A0A0P5JQA6 A0A0P5KPU8 B4MR42 B4KTX0 A0A0P5W2K4 A0A0P5F094 A0A0P5J0C0 A0A0P5HFS0 A0A0P5JAW3 A0A0N8B7R2 A0A0P5B0F4 A0A0N8BH04 A0A0P5P0K8 A0A0P5LWJ3 A0A0P5N858 A0A0P5KL43 A0A182JB17 A0A0P5H378 E9HMR4 A0A0N8BYX0 A0A226DFQ9 A0A0P5S515 A0A0P5AAG3 A0A3B3X9L6 A0A0U5GE50 E9GTS4 A0A087XBW5 A0A0P4WMI0 B4MIW8 A0A0P6D9W4 B3MF95 A0A2R8ZWR3 A0A0P5Z033 A0A0P5BPY9 E9ITE0 Q0X0F2 A0A0P5LVW7 H2NWX5 A0A0P5TVM6 A0A0P4WUK7 A0A0N8B280
A0A0A2KWH5 A0A226D5J2 A0A0M9A9Z8 A0A3D8S5S2 A0A0M5IZZ2 A0A232EXU8 E9H014 Q8MNY6 A0A068F5B8 A0A0P5XLE4 A0A3M6TQ33 A0A0P5VLQ4 A0A0N8CRT0 B4JVV3 A0A0P4YLS3 A0A0N8BDG1 A0A0N8CYB7 A0A212FL72 A0A194R423 A0A3B0JSD8 A0A0P5V5F0 A0A0P5GPW3 A0A0N8AVR2 A0A182GGY4 A0A0P5FVM8 A0A2I3SE40 A0A0P6BEL6 A0A0P5JH56 A0A0P5QQH8 A0A0P5GA08 A0A0P6AQ80 A0A1L8HMQ5 A0A0P6E6W6 G3S3C4 A0A0P5L024 A0A1W7R806 A0A0P5MDH3 Q5QBF4 A0A1I8Q0B3 A0A0P5GX76 A0A0P6F713 K7J4L1 A0A0P5J6E5 A0A0P5GGK0 A0A0P5GL06 A0A0P5FRW8 A0A0P5GQL0 A0A232FET9 F6XTG5 A0A0P5KPC1 A0A0P5G8N5 A0A0P5JQA6 A0A0P5KPU8 B4MR42 B4KTX0 A0A0P5W2K4 A0A0P5F094 A0A0P5J0C0 A0A0P5HFS0 A0A0P5JAW3 A0A0N8B7R2 A0A0P5B0F4 A0A0N8BH04 A0A0P5P0K8 A0A0P5LWJ3 A0A0P5N858 A0A0P5KL43 A0A182JB17 A0A0P5H378 E9HMR4 A0A0N8BYX0 A0A226DFQ9 A0A0P5S515 A0A0P5AAG3 A0A3B3X9L6 A0A0U5GE50 E9GTS4 A0A087XBW5 A0A0P4WMI0 B4MIW8 A0A0P6D9W4 B3MF95 A0A2R8ZWR3 A0A0P5Z033 A0A0P5BPY9 E9ITE0 Q0X0F2 A0A0P5LVW7 H2NWX5 A0A0P5TVM6 A0A0P4WUK7 A0A0N8B280
Pubmed
EMBL
BABH01010385
KQ460779
KPJ12278.1
NWSH01000531
PCG75853.1
AGBW02009839
+ More
OWR49768.1 KQ459598 KPI94661.1 JTDY01001149 KOB74739.1 JQGA01001183 KGO68710.1 LNIX01000035 OXA40118.1 KQ435706 KOX80247.1 PVWQ01000005 RDW81550.1 CP012524 ALC41574.1 NNAY01001658 OXU23296.1 GL732579 EFX74965.1 AJ316142 CAC87119.1 KJ512120 AID60343.1 GDIP01070358 JAM33357.1 RCHS01003197 RMX43535.1 GDIP01098308 JAM05407.1 GDIP01105207 JAL98507.1 CH916375 EDV98091.1 GDIP01226996 JAI96405.1 GDIQ01188571 GDIQ01097706 JAK63154.1 GDIP01086911 JAM16804.1 AGBW02007905 OWR54439.1 KPJ12274.1 OUUW01000001 SPP74008.1 GDIP01105259 JAL98455.1 GDIQ01241312 JAK10413.1 GDIQ01238163 JAK13562.1 JXUM01062824 KQ562217 KXJ76377.1 GDIQ01250854 JAK00871.1 AACZ04007130 AACZ04007131 AACZ04007132 AACZ04007133 AACZ04007134 AACZ04069008 GDIP01015982 JAM87733.1 GDIQ01202773 JAK48952.1 GDIQ01131298 JAL20428.1 GDIQ01249258 JAK02467.1 GDIP01027762 JAM75953.1 CM004467 OCT97345.1 GDIQ01066768 JAN27969.1 CABD030110710 CABD030110711 CABD030110712 GDIQ01185517 JAK66208.1 GEHC01000378 JAV47267.1 GDIQ01168977 JAK82748.1 AY752857 AAV84270.1 GDIQ01241313 JAK10412.1 GDIQ01060360 JAN34377.1 AAZX01004421 GDIQ01204564 JAK47161.1 GDIQ01241311 JAK10414.1 GDIQ01239757 JAK11968.1 GDIQ01250853 JAK00872.1 GDIQ01238164 JAK13561.1 NNAY01000351 OXU28988.1 AAMC01058510 AAMC01058511 AAMC01058512 GDIQ01188570 JAK63155.1 GDIQ01245759 JAK05966.1 GDIQ01200167 JAK51558.1 GDIQ01181367 JAK70358.1 CH963849 EDW74581.1 CH933808 EDW10696.1 GDIP01092810 JAM10905.1 GDIQ01269589 JAJ82135.1 GDIQ01204965 JAK46760.1 GDIQ01227502 JAK24223.1 GDIQ01202772 JAK48953.1 GDIQ01204563 JAK47162.1 GDIP01195844 JAJ27558.1 GDIQ01178627 JAK73098.1 GDIQ01145242 JAL06484.1 GDIQ01188569 JAK63156.1 GDIQ01167980 JAK83745.1 GDIQ01183672 JAK68053.1 GDIQ01239756 JAK11969.1 GL732689 EFX66983.1 GDIQ01131299 JAL20427.1 LNIX01000021 OXA43818.1 GDIQ01109447 JAL42279.1 GDIP01211296 GDIP01201839 GDIP01088364 JAJ21563.1 CDMC01000014 CEL09341.1 GL732564 EFX77160.1 AYCK01003435 GDIP01254171 JAI69230.1 CH963719 EDW72057.1 GDIQ01081317 JAN13420.1 CH902619 EDV35569.1 AJFE02065534 AJFE02065535 AJFE02065536 AJFE02065537 AJFE02065538 AJFE02065539 AJFE02065540 AJFE02065541 GDIP01050920 JAM52795.1 GDIP01185491 GDIP01095558 JAJ37911.1 GL765541 EFZ16163.1 AB109390 BAF02295.1 GDIQ01188871 JAK62854.1 ABGA01121506 ABGA01121507 ABGA01121508 ABGA01121509 ABGA01121510 ABGA01121511 ABGA01121512 ABGA01121513 ABGA01121514 ABGA01121515 GDIP01123509 JAL80205.1 GDIP01251974 JAI71427.1 GDIQ01220019 JAK31706.1
OWR49768.1 KQ459598 KPI94661.1 JTDY01001149 KOB74739.1 JQGA01001183 KGO68710.1 LNIX01000035 OXA40118.1 KQ435706 KOX80247.1 PVWQ01000005 RDW81550.1 CP012524 ALC41574.1 NNAY01001658 OXU23296.1 GL732579 EFX74965.1 AJ316142 CAC87119.1 KJ512120 AID60343.1 GDIP01070358 JAM33357.1 RCHS01003197 RMX43535.1 GDIP01098308 JAM05407.1 GDIP01105207 JAL98507.1 CH916375 EDV98091.1 GDIP01226996 JAI96405.1 GDIQ01188571 GDIQ01097706 JAK63154.1 GDIP01086911 JAM16804.1 AGBW02007905 OWR54439.1 KPJ12274.1 OUUW01000001 SPP74008.1 GDIP01105259 JAL98455.1 GDIQ01241312 JAK10413.1 GDIQ01238163 JAK13562.1 JXUM01062824 KQ562217 KXJ76377.1 GDIQ01250854 JAK00871.1 AACZ04007130 AACZ04007131 AACZ04007132 AACZ04007133 AACZ04007134 AACZ04069008 GDIP01015982 JAM87733.1 GDIQ01202773 JAK48952.1 GDIQ01131298 JAL20428.1 GDIQ01249258 JAK02467.1 GDIP01027762 JAM75953.1 CM004467 OCT97345.1 GDIQ01066768 JAN27969.1 CABD030110710 CABD030110711 CABD030110712 GDIQ01185517 JAK66208.1 GEHC01000378 JAV47267.1 GDIQ01168977 JAK82748.1 AY752857 AAV84270.1 GDIQ01241313 JAK10412.1 GDIQ01060360 JAN34377.1 AAZX01004421 GDIQ01204564 JAK47161.1 GDIQ01241311 JAK10414.1 GDIQ01239757 JAK11968.1 GDIQ01250853 JAK00872.1 GDIQ01238164 JAK13561.1 NNAY01000351 OXU28988.1 AAMC01058510 AAMC01058511 AAMC01058512 GDIQ01188570 JAK63155.1 GDIQ01245759 JAK05966.1 GDIQ01200167 JAK51558.1 GDIQ01181367 JAK70358.1 CH963849 EDW74581.1 CH933808 EDW10696.1 GDIP01092810 JAM10905.1 GDIQ01269589 JAJ82135.1 GDIQ01204965 JAK46760.1 GDIQ01227502 JAK24223.1 GDIQ01202772 JAK48953.1 GDIQ01204563 JAK47162.1 GDIP01195844 JAJ27558.1 GDIQ01178627 JAK73098.1 GDIQ01145242 JAL06484.1 GDIQ01188569 JAK63156.1 GDIQ01167980 JAK83745.1 GDIQ01183672 JAK68053.1 GDIQ01239756 JAK11969.1 GL732689 EFX66983.1 GDIQ01131299 JAL20427.1 LNIX01000021 OXA43818.1 GDIQ01109447 JAL42279.1 GDIP01211296 GDIP01201839 GDIP01088364 JAJ21563.1 CDMC01000014 CEL09341.1 GL732564 EFX77160.1 AYCK01003435 GDIP01254171 JAI69230.1 CH963719 EDW72057.1 GDIQ01081317 JAN13420.1 CH902619 EDV35569.1 AJFE02065534 AJFE02065535 AJFE02065536 AJFE02065537 AJFE02065538 AJFE02065539 AJFE02065540 AJFE02065541 GDIP01050920 JAM52795.1 GDIP01185491 GDIP01095558 JAJ37911.1 GL765541 EFZ16163.1 AB109390 BAF02295.1 GDIQ01188871 JAK62854.1 ABGA01121506 ABGA01121507 ABGA01121508 ABGA01121509 ABGA01121510 ABGA01121511 ABGA01121512 ABGA01121513 ABGA01121514 ABGA01121515 GDIP01123509 JAL80205.1 GDIP01251974 JAI71427.1 GDIQ01220019 JAK31706.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000053268
UP000037510
+ More
UP000030104 UP000198287 UP000053105 UP000256690 UP000092553 UP000215335 UP000000305 UP000275408 UP000001070 UP000268350 UP000069940 UP000249989 UP000002277 UP000186698 UP000001519 UP000095300 UP000002358 UP000008143 UP000007798 UP000009192 UP000075880 UP000261480 UP000054771 UP000028760 UP000007801 UP000240080 UP000001595
UP000030104 UP000198287 UP000053105 UP000256690 UP000092553 UP000215335 UP000000305 UP000275408 UP000001070 UP000268350 UP000069940 UP000249989 UP000002277 UP000186698 UP000001519 UP000095300 UP000002358 UP000008143 UP000007798 UP000009192 UP000075880 UP000261480 UP000054771 UP000028760 UP000007801 UP000240080 UP000001595
Interpro
IPR009003
Peptidase_S1_PA
+ More
IPR001314 Peptidase_S1A
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR012224 Pept_S1A_FX
IPR033116 TRYPSIN_SER
IPR009091 RCC1/BLIP-II
IPR000408 Reg_chr_condens
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR036364 SEA_dom_sf
IPR000082 SEA_dom
IPR036772 SRCR-like_dom_sf
IPR001190 SRCR
IPR017448 SRCR-like_dom
IPR001314 Peptidase_S1A
IPR001254 Trypsin_dom
IPR018114 TRYPSIN_HIS
IPR012224 Pept_S1A_FX
IPR033116 TRYPSIN_SER
IPR009091 RCC1/BLIP-II
IPR000408 Reg_chr_condens
IPR036055 LDL_receptor-like_sf
IPR002172 LDrepeatLR_classA_rpt
IPR036364 SEA_dom_sf
IPR000082 SEA_dom
IPR036772 SRCR-like_dom_sf
IPR001190 SRCR
IPR017448 SRCR-like_dom
SUPFAM
Gene 3D
ProteinModelPortal
H9J0I8
A0A194R398
A0A2A4JWI7
A0A212F7S3
A0A194PMK0
A0A0L7LH96
+ More
A0A0A2KWH5 A0A226D5J2 A0A0M9A9Z8 A0A3D8S5S2 A0A0M5IZZ2 A0A232EXU8 E9H014 Q8MNY6 A0A068F5B8 A0A0P5XLE4 A0A3M6TQ33 A0A0P5VLQ4 A0A0N8CRT0 B4JVV3 A0A0P4YLS3 A0A0N8BDG1 A0A0N8CYB7 A0A212FL72 A0A194R423 A0A3B0JSD8 A0A0P5V5F0 A0A0P5GPW3 A0A0N8AVR2 A0A182GGY4 A0A0P5FVM8 A0A2I3SE40 A0A0P6BEL6 A0A0P5JH56 A0A0P5QQH8 A0A0P5GA08 A0A0P6AQ80 A0A1L8HMQ5 A0A0P6E6W6 G3S3C4 A0A0P5L024 A0A1W7R806 A0A0P5MDH3 Q5QBF4 A0A1I8Q0B3 A0A0P5GX76 A0A0P6F713 K7J4L1 A0A0P5J6E5 A0A0P5GGK0 A0A0P5GL06 A0A0P5FRW8 A0A0P5GQL0 A0A232FET9 F6XTG5 A0A0P5KPC1 A0A0P5G8N5 A0A0P5JQA6 A0A0P5KPU8 B4MR42 B4KTX0 A0A0P5W2K4 A0A0P5F094 A0A0P5J0C0 A0A0P5HFS0 A0A0P5JAW3 A0A0N8B7R2 A0A0P5B0F4 A0A0N8BH04 A0A0P5P0K8 A0A0P5LWJ3 A0A0P5N858 A0A0P5KL43 A0A182JB17 A0A0P5H378 E9HMR4 A0A0N8BYX0 A0A226DFQ9 A0A0P5S515 A0A0P5AAG3 A0A3B3X9L6 A0A0U5GE50 E9GTS4 A0A087XBW5 A0A0P4WMI0 B4MIW8 A0A0P6D9W4 B3MF95 A0A2R8ZWR3 A0A0P5Z033 A0A0P5BPY9 E9ITE0 Q0X0F2 A0A0P5LVW7 H2NWX5 A0A0P5TVM6 A0A0P4WUK7 A0A0N8B280
A0A0A2KWH5 A0A226D5J2 A0A0M9A9Z8 A0A3D8S5S2 A0A0M5IZZ2 A0A232EXU8 E9H014 Q8MNY6 A0A068F5B8 A0A0P5XLE4 A0A3M6TQ33 A0A0P5VLQ4 A0A0N8CRT0 B4JVV3 A0A0P4YLS3 A0A0N8BDG1 A0A0N8CYB7 A0A212FL72 A0A194R423 A0A3B0JSD8 A0A0P5V5F0 A0A0P5GPW3 A0A0N8AVR2 A0A182GGY4 A0A0P5FVM8 A0A2I3SE40 A0A0P6BEL6 A0A0P5JH56 A0A0P5QQH8 A0A0P5GA08 A0A0P6AQ80 A0A1L8HMQ5 A0A0P6E6W6 G3S3C4 A0A0P5L024 A0A1W7R806 A0A0P5MDH3 Q5QBF4 A0A1I8Q0B3 A0A0P5GX76 A0A0P6F713 K7J4L1 A0A0P5J6E5 A0A0P5GGK0 A0A0P5GL06 A0A0P5FRW8 A0A0P5GQL0 A0A232FET9 F6XTG5 A0A0P5KPC1 A0A0P5G8N5 A0A0P5JQA6 A0A0P5KPU8 B4MR42 B4KTX0 A0A0P5W2K4 A0A0P5F094 A0A0P5J0C0 A0A0P5HFS0 A0A0P5JAW3 A0A0N8B7R2 A0A0P5B0F4 A0A0N8BH04 A0A0P5P0K8 A0A0P5LWJ3 A0A0P5N858 A0A0P5KL43 A0A182JB17 A0A0P5H378 E9HMR4 A0A0N8BYX0 A0A226DFQ9 A0A0P5S515 A0A0P5AAG3 A0A3B3X9L6 A0A0U5GE50 E9GTS4 A0A087XBW5 A0A0P4WMI0 B4MIW8 A0A0P6D9W4 B3MF95 A0A2R8ZWR3 A0A0P5Z033 A0A0P5BPY9 E9ITE0 Q0X0F2 A0A0P5LVW7 H2NWX5 A0A0P5TVM6 A0A0P4WUK7 A0A0N8B280
PDB
1EQ9
E-value=3.59579e-24,
Score=274
Ontologies
GO
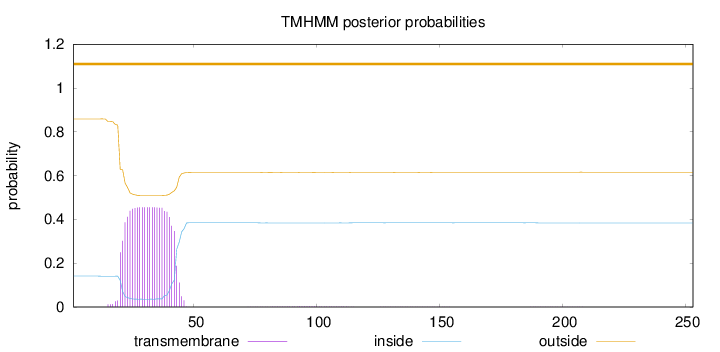
Topology
Length:
253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.2626
Exp number, first 60 AAs:
10.15778
Total prob of N-in:
0.14101
POSSIBLE N-term signal
sequence
outside
1 - 253
Population Genetic Test Statistics
Pi
148.219353
Theta
75.482949
Tajima's D
-0.541317
CLR
29.718684
CSRT
0.231988400579971
Interpretation
Uncertain
