Gene
KWMTBOMO02174
Pre Gene Modal
BGIBMGA003024
Annotation
PREDICTED:_visual_pigment-like_receptor_peropsin_[Amyelois_transitella]
Full name
Visual pigment-like receptor peropsin
Location in the cell
PlasmaMembrane Reliability : 4.976
Sequence
CDS
ATGGGAGTGTTTGGGGCGCTGCTTAACGGCTGGTTGTTCGCTACCTTTATTCATAGCCACTTGTTAATTTTCAAAAGTCACATATTGGTATTCAATCTGTGCGCGGCATCGCTCGGAAGGAATTTACTGGGATTTCCTTTCTCGGGTTCTTCAGCTGTAGCAAAAAGATGGCTGTTCGGTCCATCCTGTTGTCAGCTATTTGCGTTTTTCAATCAATTCTTTGGTGTCTTTCAAATGACTTCAATATTCGTTTTAATATTGGAACGATATATTCAAGTTAAGTTTTATAAGCGTGAAAAATCGGTGTACATAAGGTTGTATTGGACACTGATCGCAGTAAGTTGGATCAATTCATTTCTATTCGCCACTCCTCCTCTGTTCGGATACGGATTATATTCGTGTGACACAACTGGAACATCATGCACCTTTCTTTGGCCTTCAATGTCATCGGGAGCAAAGCAATTGGGTTACGCCGTGCCCTACATTTTAATTTGTGGACTCATACCAATTGTGGCAATGTTTTACTACATGGGAAAAGCGTTAAGATTAGAAAAGATATTTTACAAGGGTGAACAACAAAGGGAACAAAAATGTCTTACCCAGAGCGCACACGCGGTTTGTGTTGGCACATTGGCCTTATGGATTCCAGCAGCTGTCCTAGCGGGATGGCAATGGGTACCGCTCCTCATCAAAGGCTATCGACCGCACGTGCCACCGACACTAGCTTTGATCGCTTCCATTGCTTCTGAAGCAGCAACTACTGTTCCTGTGCTGTGTTACCTAGCCGTGGATGAGAGACTCAGAGCAGCGTTGTTGGGTAGAATGCGAAAACAATACGCTTTACTTCAACCCGAGCGAGCAAAGATTTATAAGAGAGTATAG
Protein
MGVFGALLNGWLFATFIHSHLLIFKSHILVFNLCAASLGRNLLGFPFSGSSAVAKRWLFGPSCCQLFAFFNQFFGVFQMTSIFVLILERYIQVKFYKREKSVYIRLYWTLIAVSWINSFLFATPPLFGYGLYSCDTTGTSCTFLWPSMSSGAKQLGYAVPYILICGLIPIVAMFYYMGKALRLEKIFYKGEQQREQKCLTQSAHAVCVGTLALWIPAAVLAGWQWVPLLIKGYRPHVPPTLALIASIASEAATTVPVLCYLAVDERLRAALLGRMRKQYALLQPERAKIYKRV
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family. Opsin subfamily.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the G-protein coupled receptor 2 family.
Belongs to the G-protein coupled receptor 1 family.
Belongs to the G-protein coupled receptor 2 family.
Keywords
Chromophore
Complete proteome
Disulfide bond
G-protein coupled receptor
Glycoprotein
Membrane
Photoreceptor protein
Receptor
Reference proteome
Retinal protein
Sensory transduction
Transducer
Transmembrane
Transmembrane helix
Feature
chain Visual pigment-like receptor peropsin
Uniprot
A0A2A4JSF6
A0A194PPK4
A0A194R3L0
A0A1B6MMX8
A0A1B6K4H0
A0A0C6G2Z6
+ More
A0A0C6FZD7 A0A0C6FZG7 E1CFG1 A0A0C6G6P8 A0A0C6FZ99 A0A0C6FZB7 A0A0C6FZF7 A0A0C6FPQ1 A0A0C6FPS0 A0A0C6FPG7 A0A0C6G6P3 A0A0C9LM67 A0A0C6FPI7 A0A0G3Y5J3 A0A1L8F800 A0A1B6ET89 F7BWP4 C3XR28 A0A1S3JHZ2 A0A151M623 A0A142BLT2 A0A0P0YPZ8 A0A2H4KKR6 A0A0H2UI65 A0A3N0Z0K2 A0A3B4DM11 Q543W9 O35214 A0A1L8GAX0 A0A1S3FT77 R9R6L5 A0A2R8Q4C0 A0A182HT24 A0A1L8F3C7 A0A2S2QM89 W5LC48 A0A1W2Q6H2 J9K8Z3 A0A2Z4C5M8 Q17A90 A0A2U8T683 A0A1A8LM78 A0A3B4B3G8 A0A094LU95 A0A2S2PM78 F6UZB2 A0A1S4FAC5 E0R7P4 G3T3L1 A0A1A8NTU5 A0A1V9XRA4 A0A3B5B3N3 T2I4T4 A0A3P8WFL4 A0A182XWZ2 A0A1S3RSC0 A0A2H8TRG1 W5N199 A0A2I4AXG3 A0A182F8X5 A0A3B3D7R5 A0A0F8BCQ8
A0A0C6FZD7 A0A0C6FZG7 E1CFG1 A0A0C6G6P8 A0A0C6FZ99 A0A0C6FZB7 A0A0C6FZF7 A0A0C6FPQ1 A0A0C6FPS0 A0A0C6FPG7 A0A0C6G6P3 A0A0C9LM67 A0A0C6FPI7 A0A0G3Y5J3 A0A1L8F800 A0A1B6ET89 F7BWP4 C3XR28 A0A1S3JHZ2 A0A151M623 A0A142BLT2 A0A0P0YPZ8 A0A2H4KKR6 A0A0H2UI65 A0A3N0Z0K2 A0A3B4DM11 Q543W9 O35214 A0A1L8GAX0 A0A1S3FT77 R9R6L5 A0A2R8Q4C0 A0A182HT24 A0A1L8F3C7 A0A2S2QM89 W5LC48 A0A1W2Q6H2 J9K8Z3 A0A2Z4C5M8 Q17A90 A0A2U8T683 A0A1A8LM78 A0A3B4B3G8 A0A094LU95 A0A2S2PM78 F6UZB2 A0A1S4FAC5 E0R7P4 G3T3L1 A0A1A8NTU5 A0A1V9XRA4 A0A3B5B3N3 T2I4T4 A0A3P8WFL4 A0A182XWZ2 A0A1S3RSC0 A0A2H8TRG1 W5N199 A0A2I4AXG3 A0A182F8X5 A0A3B3D7R5 A0A0F8BCQ8
Pubmed
EMBL
NWSH01000765
PCG74332.1
KQ459598
KPI94664.1
KQ460779
KPJ12282.1
+ More
GEBQ01002669 JAT37308.1 GECU01001368 JAT06339.1 LC009259 BAQ54907.1 LC009236 BAQ54883.1 LC009290 BAQ54938.1 AB525082 BAJ22674.1 LC009122 BAQ54765.1 LC009167 BAQ54813.1 LC009202 BAQ54848.1 LC009275 BAQ54923.1 LC009147 BAQ54791.1 LC009185 BAQ54831.1 LC009058 BAQ54696.1 LC009098 BAQ54740.1 BR001239 FAA01168.1 LC009078 BAQ54716.1 KP725074 AKM21242.1 CM004480 OCT67723.1 GECZ01028668 JAS41101.1 AAMC01014454 AAMC01014455 GG666456 EEN69146.1 AKHW03006480 KYO19967.1 KU682725 AMP19653.1 LC009375 BAT23016.1 MF489903 ASW31197.1 ACPB03017884 RJVU01018281 ROL51743.1 AK044717 CH466532 BAC32047.1 EDL12234.1 AF012271 CM004474 OCT81067.1 CR352209 CU302291 JX293360 KT008412 AGK24996.1 ALG92552.1 APCN01000400 APCN01000401 CM004481 OCT66080.1 GGMS01009641 MBY78844.1 AC117142 ABLF02037968 ABLF02037969 ABLF02037974 ABLF02037978 ABLF02037979 ABLF02037980 ABLF02037981 ABLF02037982 ABLF02037985 MF464468 AWU78798.1 CH477338 EAT43163.2 MF434471 MF434472 AWM72030.1 AWM72031.1 HAEF01007935 SBR45648.1 KL278746 KFZ67109.1 GGMR01017951 MBY30570.1 AAMC01052310 AAMC01052311 AAMC01052312 AAMC01052313 AAMC01052314 AY377392 AAR24539.1 OCT81316.1 HAEH01003891 SBR72480.1 MNPL01005422 OQR76027.1 HF566406 CCP46949.1 GFXV01004979 MBW16784.1 AHAT01017896 KQ041021 KKF31418.1
GEBQ01002669 JAT37308.1 GECU01001368 JAT06339.1 LC009259 BAQ54907.1 LC009236 BAQ54883.1 LC009290 BAQ54938.1 AB525082 BAJ22674.1 LC009122 BAQ54765.1 LC009167 BAQ54813.1 LC009202 BAQ54848.1 LC009275 BAQ54923.1 LC009147 BAQ54791.1 LC009185 BAQ54831.1 LC009058 BAQ54696.1 LC009098 BAQ54740.1 BR001239 FAA01168.1 LC009078 BAQ54716.1 KP725074 AKM21242.1 CM004480 OCT67723.1 GECZ01028668 JAS41101.1 AAMC01014454 AAMC01014455 GG666456 EEN69146.1 AKHW03006480 KYO19967.1 KU682725 AMP19653.1 LC009375 BAT23016.1 MF489903 ASW31197.1 ACPB03017884 RJVU01018281 ROL51743.1 AK044717 CH466532 BAC32047.1 EDL12234.1 AF012271 CM004474 OCT81067.1 CR352209 CU302291 JX293360 KT008412 AGK24996.1 ALG92552.1 APCN01000400 APCN01000401 CM004481 OCT66080.1 GGMS01009641 MBY78844.1 AC117142 ABLF02037968 ABLF02037969 ABLF02037974 ABLF02037978 ABLF02037979 ABLF02037980 ABLF02037981 ABLF02037982 ABLF02037985 MF464468 AWU78798.1 CH477338 EAT43163.2 MF434471 MF434472 AWM72030.1 AWM72031.1 HAEF01007935 SBR45648.1 KL278746 KFZ67109.1 GGMR01017951 MBY30570.1 AAMC01052310 AAMC01052311 AAMC01052312 AAMC01052313 AAMC01052314 AY377392 AAR24539.1 OCT81316.1 HAEH01003891 SBR72480.1 MNPL01005422 OQR76027.1 HF566406 CCP46949.1 GFXV01004979 MBW16784.1 AHAT01017896 KQ041021 KKF31418.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000186698
UP000008143
UP000001554
+ More
UP000085678 UP000050525 UP000015103 UP000261440 UP000000589 UP000081671 UP000000437 UP000075840 UP000018467 UP000002494 UP000007819 UP000008820 UP000261520 UP000007646 UP000192247 UP000261400 UP000265120 UP000076408 UP000087266 UP000018468 UP000192220 UP000069272 UP000261560
UP000085678 UP000050525 UP000015103 UP000261440 UP000000589 UP000081671 UP000000437 UP000075840 UP000018467 UP000002494 UP000007819 UP000008820 UP000261520 UP000007646 UP000192247 UP000261400 UP000265120 UP000076408 UP000087266 UP000018468 UP000192220 UP000069272 UP000261560
PRIDE
Interpro
SUPFAM
SSF49899
SSF49899
ProteinModelPortal
A0A2A4JSF6
A0A194PPK4
A0A194R3L0
A0A1B6MMX8
A0A1B6K4H0
A0A0C6G2Z6
+ More
A0A0C6FZD7 A0A0C6FZG7 E1CFG1 A0A0C6G6P8 A0A0C6FZ99 A0A0C6FZB7 A0A0C6FZF7 A0A0C6FPQ1 A0A0C6FPS0 A0A0C6FPG7 A0A0C6G6P3 A0A0C9LM67 A0A0C6FPI7 A0A0G3Y5J3 A0A1L8F800 A0A1B6ET89 F7BWP4 C3XR28 A0A1S3JHZ2 A0A151M623 A0A142BLT2 A0A0P0YPZ8 A0A2H4KKR6 A0A0H2UI65 A0A3N0Z0K2 A0A3B4DM11 Q543W9 O35214 A0A1L8GAX0 A0A1S3FT77 R9R6L5 A0A2R8Q4C0 A0A182HT24 A0A1L8F3C7 A0A2S2QM89 W5LC48 A0A1W2Q6H2 J9K8Z3 A0A2Z4C5M8 Q17A90 A0A2U8T683 A0A1A8LM78 A0A3B4B3G8 A0A094LU95 A0A2S2PM78 F6UZB2 A0A1S4FAC5 E0R7P4 G3T3L1 A0A1A8NTU5 A0A1V9XRA4 A0A3B5B3N3 T2I4T4 A0A3P8WFL4 A0A182XWZ2 A0A1S3RSC0 A0A2H8TRG1 W5N199 A0A2I4AXG3 A0A182F8X5 A0A3B3D7R5 A0A0F8BCQ8
A0A0C6FZD7 A0A0C6FZG7 E1CFG1 A0A0C6G6P8 A0A0C6FZ99 A0A0C6FZB7 A0A0C6FZF7 A0A0C6FPQ1 A0A0C6FPS0 A0A0C6FPG7 A0A0C6G6P3 A0A0C9LM67 A0A0C6FPI7 A0A0G3Y5J3 A0A1L8F800 A0A1B6ET89 F7BWP4 C3XR28 A0A1S3JHZ2 A0A151M623 A0A142BLT2 A0A0P0YPZ8 A0A2H4KKR6 A0A0H2UI65 A0A3N0Z0K2 A0A3B4DM11 Q543W9 O35214 A0A1L8GAX0 A0A1S3FT77 R9R6L5 A0A2R8Q4C0 A0A182HT24 A0A1L8F3C7 A0A2S2QM89 W5LC48 A0A1W2Q6H2 J9K8Z3 A0A2Z4C5M8 Q17A90 A0A2U8T683 A0A1A8LM78 A0A3B4B3G8 A0A094LU95 A0A2S2PM78 F6UZB2 A0A1S4FAC5 E0R7P4 G3T3L1 A0A1A8NTU5 A0A1V9XRA4 A0A3B5B3N3 T2I4T4 A0A3P8WFL4 A0A182XWZ2 A0A1S3RSC0 A0A2H8TRG1 W5N199 A0A2I4AXG3 A0A182F8X5 A0A3B3D7R5 A0A0F8BCQ8
PDB
5WKT
E-value=1.56154e-05,
Score=114
Ontologies
GO
PANTHER
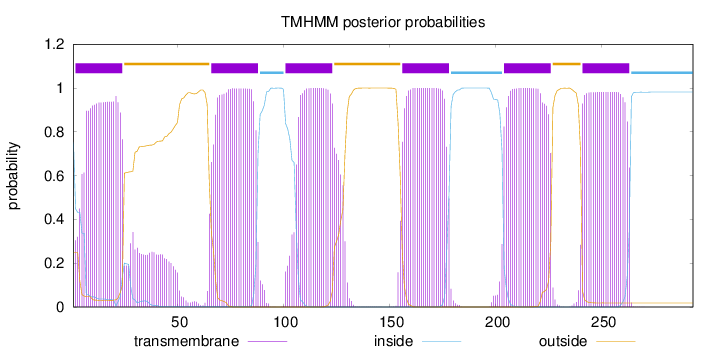
Topology
Subcellular location
Membrane
Length:
293
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
137.02627
Exp number, first 60 AAs:
25.12857
Total prob of N-in:
0.75237
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 24
outside
25 - 65
TMhelix
66 - 88
inside
89 - 100
TMhelix
101 - 123
outside
124 - 155
TMhelix
156 - 178
inside
179 - 203
TMhelix
204 - 226
outside
227 - 240
TMhelix
241 - 263
inside
264 - 293
Population Genetic Test Statistics
Pi
398.777841
Theta
225.137687
Tajima's D
2.428743
CLR
0.011448
CSRT
0.936003199840008
Interpretation
Uncertain
