Gene
KWMTBOMO02173
Pre Gene Modal
BGIBMGA003139
Annotation
PREDICTED:_protein_msta?_isoform_A-like_isoform_X1_[Bombyx_mori]
Full name
SET domain-containing protein SmydA-8, isoform A
+ More
SET domain-containing protein SmydA-8, isoform B
SET domain-containing protein SmydA-8, isoform B
Location in the cell
PlasmaMembrane Reliability : 2.035
Sequence
CDS
ATGCTATCATCAAACCAATTGACGCAGATAGTGCACAAATATCTCGGTCTCTATAACTCTACAGACTGCATACAGCAATTCTGCAAATGGACGGTCCGAGAATCAACGGTGGGCGGTCGAGGAATAATAGCCACTGAAGACATTCCGTGTGGAGAAGTTTTGTTTGTTGATCGTCCTTTGATTTTTGGTCCGCGTTGTGGAAATGTCATCAGAGGATGTACAGTTTGCTCCAAATTCGAAGATGACGGATTCTTTAAGTGTTCTCTGTGCGCCCTATTGCTATGCTCAACGGAATGTCAAGATTTTCACACTGATGATTGCAACATTATTGCTGGTTGGAAGAAAAAAGTTGACATTGAAGAAATAGACGAAATTGTAATGTCACATGCCCTGGCCGCCATAAGAGCATTGTCTTTAAACCAAGAACAAAAACAATTTATGAGCGCTTTAGCAGCACACACATTACCACAACATGGCGGTGAAATCATAAAACTCAAGCAGTATTTTGAAATACCTGAAGAAATTGAAAAAGATATGATATTCGCTTGTCAAGCTTTAGATACAAATGCATTTCAAATTGCAACACCGTACGGCAAGAAGGAAATGAGTCTTCGTGGTTTGTACCCTGTAGCAGGACTCATGAATCATAGTTGTTTGCCAAATGCGACATATAGTTACAACGATGAACGTCAAATGATCGTTAAAACGAGTCGATTCATTGCAGCTGGAACTGAAATATTCACTTGCTATTCTGGGATTTTATGGGGAACGACGACTCGACGCAGTTATTTATTCAAAACAAAACATTTTTGGTGCAAGTGTGAACGATGCGCGGATCCTACGGAAAACGGTACTATGCTCGCTGCCCTCTGTTGTTTAGCGAACGAATGTACGACGGGATATCTTCTGCCGATCGAACCACTGAAGTCTTCGTCTTCATGGCAATGTACAGCGTGTGGTCTCCGCGTGCCAAGTAAAATTATCTACAAAATACAGGTAGCTTTAGGGACCTTAATAAGCACTTTAGACTTCCAAAACCTTGAACAAATGGAGAATTTCCTAATGAAAAGGGTCACAAAATTCGTGCCAAAAGTTAATCACATAGTCGTCGACCTCCAATGCCGACTCATATGGGAATTCGGAGACGTACAAGGGTATCGCTGGCACGAGCTAACGGAATCGCGGCTATCCCTGAAATTGGTCCTGTGCCGAGATATATTGGCCACGCTGGCGGCCCTGGGTTTAGAAGACTCGCACCTGTGCGGCCTGCTATTCTACCACCTGCACGCCACGCTAGCAGAACGCGCCAGGCGCCACCCAGACTTATATGACGGTCTGAAATCAGAAATAGAGAGTACCATAGAAAGAGCGTACCGAATCCTCCGAGGCGATATATCCGCTCCAGTTGACCTTGAGTTGAGATATCGCTATCTTGGTCCCGATTGTGAAAAACCACAGCAAGAGAGATTCTCCATCCTCAGTATATAA
Protein
MLSSNQLTQIVHKYLGLYNSTDCIQQFCKWTVRESTVGGRGIIATEDIPCGEVLFVDRPLIFGPRCGNVIRGCTVCSKFEDDGFFKCSLCALLLCSTECQDFHTDDCNIIAGWKKKVDIEEIDEIVMSHALAAIRALSLNQEQKQFMSALAAHTLPQHGGEIIKLKQYFEIPEEIEKDMIFACQALDTNAFQIATPYGKKEMSLRGLYPVAGLMNHSCLPNATYSYNDERQMIVKTSRFIAAGTEIFTCYSGILWGTTTRRSYLFKTKHFWCKCERCADPTENGTMLAALCCLANECTTGYLLPIEPLKSSSSWQCTACGLRVPSKIIYKIQVALGTLISTLDFQNLEQMENFLMKRVTKFVPKVNHIVVDLQCRLIWEFGDVQGYRWHELTESRLSLKLVLCRDILATLAALGLEDSHLCGLLFYHLHATLAERARRHPDLYDGLKSEIESTIERAYRILRGDISAPVDLELRYRYLGPDCEKPQQERFSILSI
Summary
Similarity
Belongs to the class V-like SAM-binding methyltransferase superfamily.
Keywords
Alternative splicing
Complete proteome
Methyltransferase
Reference proteome
S-adenosyl-L-methionine
Transferase
Feature
chain SET domain-containing protein SmydA-8, isoform A
Uniprot
H9J0V2
A0A2A4JRU9
A0A194PU53
A0A194R3A3
A0A2H1WK83
A0A0L7LAT1
+ More
A0A2H1WJV1 A0A2A4JQL2 D6WLG5 E2APG8 A0A1Y1MC08 A0A1Y1MDX9 A0A1Y1MF86 E9J2E4 A0A195B1H2 A0A195DJ28 A0A154P0F2 A0A3L8DDT9 A0A158NDG8 A0A195FI27 A0A151X0G6 A0A0N0U4Z4 A0A088AD93 A0A2A3ECJ9 A0A2P8ZCM0 A0A067QYV6 A0A1B6D0F2 A0A1B6E7R7 A0A0L7RA61 U4UGP7 A0A1B0CSC2 A0A1W4VT46 A0A1A9X4K0 A0A026WKD8 X2JDK3 O46040 A0A1A9XKB7 A0A1A9UU31 B5RJI3 B3MYS6 A0A1A9ZB74 B3P8Y6 A0A0A1XAS4 W8BTV1 A0A0Q5SS11 A0A1I8PSS9 A0A0K8V843 A0A1I8PSQ9 A0A0R3NXV9 A0A1I8PSV0 B4L5T8 A0A1B6C5C4 A0A1I8N6L9 B4NQ06 A0A1B0FK75 B4H372 A0A1Q3FS50 A0A1S3CTS0 A0A1J1IR78 A0A3Q0IJ68 B0XG67 T1PEZ7 A0A1I8N6M1 B5DKF2 A0A182FML8 A0A034W4S6 A0A182GQP9 A0A1W4VTU3 A0A0M3QZK5 A0A0Q5SY93 A0A1I8PSQ3 B4M7C7 P83501 W8BNI2 W5JT12 J9LMV0 A0A182PNS3 A0A0A1XGI4 B4L398 A0A0K8UI74 A0A3B0J935 A0A182MJM8 A0A182IZV8 A0A1I8N6M0 A0A1I8N6L8 A0A0A1WXV0 Q16ZX7 A0A1S4FI78 A0A1B0CEQ0 B4M3S5
A0A2H1WJV1 A0A2A4JQL2 D6WLG5 E2APG8 A0A1Y1MC08 A0A1Y1MDX9 A0A1Y1MF86 E9J2E4 A0A195B1H2 A0A195DJ28 A0A154P0F2 A0A3L8DDT9 A0A158NDG8 A0A195FI27 A0A151X0G6 A0A0N0U4Z4 A0A088AD93 A0A2A3ECJ9 A0A2P8ZCM0 A0A067QYV6 A0A1B6D0F2 A0A1B6E7R7 A0A0L7RA61 U4UGP7 A0A1B0CSC2 A0A1W4VT46 A0A1A9X4K0 A0A026WKD8 X2JDK3 O46040 A0A1A9XKB7 A0A1A9UU31 B5RJI3 B3MYS6 A0A1A9ZB74 B3P8Y6 A0A0A1XAS4 W8BTV1 A0A0Q5SS11 A0A1I8PSS9 A0A0K8V843 A0A1I8PSQ9 A0A0R3NXV9 A0A1I8PSV0 B4L5T8 A0A1B6C5C4 A0A1I8N6L9 B4NQ06 A0A1B0FK75 B4H372 A0A1Q3FS50 A0A1S3CTS0 A0A1J1IR78 A0A3Q0IJ68 B0XG67 T1PEZ7 A0A1I8N6M1 B5DKF2 A0A182FML8 A0A034W4S6 A0A182GQP9 A0A1W4VTU3 A0A0M3QZK5 A0A0Q5SY93 A0A1I8PSQ3 B4M7C7 P83501 W8BNI2 W5JT12 J9LMV0 A0A182PNS3 A0A0A1XGI4 B4L398 A0A0K8UI74 A0A3B0J935 A0A182MJM8 A0A182IZV8 A0A1I8N6M0 A0A1I8N6L8 A0A0A1WXV0 Q16ZX7 A0A1S4FI78 A0A1B0CEQ0 B4M3S5
EC Number
2.1.1.-
Pubmed
19121390
26354079
26227816
18362917
19820115
20798317
+ More
28004739 21282665 30249741 21347285 29403074 24845553 23537049 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10779958 10731137 17994087 25830018 24495485 15632085 25315136 25348373 26483478 20920257 23761445 17510324
28004739 21282665 30249741 21347285 29403074 24845553 23537049 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10779958 10731137 17994087 25830018 24495485 15632085 25315136 25348373 26483478 20920257 23761445 17510324
EMBL
BABH01010388
NWSH01000765
PCG74344.1
KQ459598
KPI94665.1
KQ460779
+ More
KPJ12283.1 ODYU01009118 SOQ53312.1 JTDY01001935 KOB72509.1 ODYU01009145 SOQ53363.1 PCG74345.1 KQ971343 EFA04109.2 GL441542 EFN64667.1 GEZM01035428 GEZM01035426 GEZM01035424 GEZM01035417 JAV83234.1 GEZM01035436 GEZM01035435 GEZM01035433 GEZM01035418 GEZM01035416 JAV83218.1 GEZM01035434 GEZM01035432 GEZM01035431 GEZM01035430 GEZM01035429 GEZM01035427 GEZM01035425 GEZM01035423 GEZM01035422 GEZM01035421 GEZM01035420 GEZM01035419 GEZM01035415 GEZM01035414 GEZM01035413 JAV83240.1 GL767845 EFZ13004.1 KQ976690 KYM78049.1 KQ980800 KYN12885.1 KQ434787 KZC05302.1 QOIP01000009 RLU18491.1 ADTU01012513 ADTU01012514 KQ981560 KYN39922.1 KQ982626 KYQ53461.1 KQ435811 KOX72882.1 KZ288285 PBC29465.1 PYGN01000104 PSN54241.1 KK853182 KDR10160.1 GEDC01018154 JAS19144.1 GEDC01003404 JAS33894.1 KQ414620 KOC67641.1 KB632288 ERL91523.1 AJWK01025894 KK107199 EZA55564.1 AE014298 AHN59283.1 AJ238707 AL009193 AAN09072.1 BT044457 ACH92522.1 CH902632 EDV32770.1 CH954183 EDV45591.1 GBXI01005903 JAD08389.1 GAMC01006167 JAC00389.1 KQS26034.1 GDHF01017386 JAI34928.1 CH379063 KRT05812.1 CH933811 EDW06547.1 GEDC01028611 JAS08687.1 CH964291 EDW86231.2 CCAG010004881 CH479205 EDW30765.1 GFDL01004614 JAV30431.1 CVRI01000057 CRL02052.1 DS232996 EDS27285.1 KA647264 AFP61893.1 EDY72001.2 GAKP01008351 JAC50601.1 JXUM01081058 JXUM01081059 JXUM01081060 JXUM01081061 KQ563224 KXJ74253.1 CP012528 ALC49539.1 KQS26033.1 CH940653 EDW62694.2 GAMC01006168 JAC00388.1 ADMH02000168 ETN67517.1 ABLF02029157 GBXI01003793 JAD10499.1 CH933810 EDW07026.1 GDHF01033587 GDHF01026086 JAI18727.1 JAI26228.1 OUUW01000003 SPP78717.1 AXCM01006441 GBXI01010408 JAD03884.1 CH477481 EAT40226.1 AJWK01009066 CH940651 EDW65450.1
KPJ12283.1 ODYU01009118 SOQ53312.1 JTDY01001935 KOB72509.1 ODYU01009145 SOQ53363.1 PCG74345.1 KQ971343 EFA04109.2 GL441542 EFN64667.1 GEZM01035428 GEZM01035426 GEZM01035424 GEZM01035417 JAV83234.1 GEZM01035436 GEZM01035435 GEZM01035433 GEZM01035418 GEZM01035416 JAV83218.1 GEZM01035434 GEZM01035432 GEZM01035431 GEZM01035430 GEZM01035429 GEZM01035427 GEZM01035425 GEZM01035423 GEZM01035422 GEZM01035421 GEZM01035420 GEZM01035419 GEZM01035415 GEZM01035414 GEZM01035413 JAV83240.1 GL767845 EFZ13004.1 KQ976690 KYM78049.1 KQ980800 KYN12885.1 KQ434787 KZC05302.1 QOIP01000009 RLU18491.1 ADTU01012513 ADTU01012514 KQ981560 KYN39922.1 KQ982626 KYQ53461.1 KQ435811 KOX72882.1 KZ288285 PBC29465.1 PYGN01000104 PSN54241.1 KK853182 KDR10160.1 GEDC01018154 JAS19144.1 GEDC01003404 JAS33894.1 KQ414620 KOC67641.1 KB632288 ERL91523.1 AJWK01025894 KK107199 EZA55564.1 AE014298 AHN59283.1 AJ238707 AL009193 AAN09072.1 BT044457 ACH92522.1 CH902632 EDV32770.1 CH954183 EDV45591.1 GBXI01005903 JAD08389.1 GAMC01006167 JAC00389.1 KQS26034.1 GDHF01017386 JAI34928.1 CH379063 KRT05812.1 CH933811 EDW06547.1 GEDC01028611 JAS08687.1 CH964291 EDW86231.2 CCAG010004881 CH479205 EDW30765.1 GFDL01004614 JAV30431.1 CVRI01000057 CRL02052.1 DS232996 EDS27285.1 KA647264 AFP61893.1 EDY72001.2 GAKP01008351 JAC50601.1 JXUM01081058 JXUM01081059 JXUM01081060 JXUM01081061 KQ563224 KXJ74253.1 CP012528 ALC49539.1 KQS26033.1 CH940653 EDW62694.2 GAMC01006168 JAC00388.1 ADMH02000168 ETN67517.1 ABLF02029157 GBXI01003793 JAD10499.1 CH933810 EDW07026.1 GDHF01033587 GDHF01026086 JAI18727.1 JAI26228.1 OUUW01000003 SPP78717.1 AXCM01006441 GBXI01010408 JAD03884.1 CH477481 EAT40226.1 AJWK01009066 CH940651 EDW65450.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007266
+ More
UP000000311 UP000078540 UP000078492 UP000076502 UP000279307 UP000005205 UP000078541 UP000075809 UP000053105 UP000005203 UP000242457 UP000245037 UP000027135 UP000053825 UP000030742 UP000092461 UP000192221 UP000091820 UP000053097 UP000000803 UP000092443 UP000078200 UP000007801 UP000092445 UP000008711 UP000095300 UP000001819 UP000009192 UP000095301 UP000007798 UP000092444 UP000008744 UP000079169 UP000183832 UP000002320 UP000069272 UP000069940 UP000249989 UP000092553 UP000008792 UP000000673 UP000007819 UP000075885 UP000268350 UP000075883 UP000075880 UP000008820
UP000000311 UP000078540 UP000078492 UP000076502 UP000279307 UP000005205 UP000078541 UP000075809 UP000053105 UP000005203 UP000242457 UP000245037 UP000027135 UP000053825 UP000030742 UP000092461 UP000192221 UP000091820 UP000053097 UP000000803 UP000092443 UP000078200 UP000007801 UP000092445 UP000008711 UP000095300 UP000001819 UP000009192 UP000095301 UP000007798 UP000092444 UP000008744 UP000079169 UP000183832 UP000002320 UP000069272 UP000069940 UP000249989 UP000092553 UP000008792 UP000000673 UP000007819 UP000075885 UP000268350 UP000075883 UP000075880 UP000008820
Gene 3D
ProteinModelPortal
H9J0V2
A0A2A4JRU9
A0A194PU53
A0A194R3A3
A0A2H1WK83
A0A0L7LAT1
+ More
A0A2H1WJV1 A0A2A4JQL2 D6WLG5 E2APG8 A0A1Y1MC08 A0A1Y1MDX9 A0A1Y1MF86 E9J2E4 A0A195B1H2 A0A195DJ28 A0A154P0F2 A0A3L8DDT9 A0A158NDG8 A0A195FI27 A0A151X0G6 A0A0N0U4Z4 A0A088AD93 A0A2A3ECJ9 A0A2P8ZCM0 A0A067QYV6 A0A1B6D0F2 A0A1B6E7R7 A0A0L7RA61 U4UGP7 A0A1B0CSC2 A0A1W4VT46 A0A1A9X4K0 A0A026WKD8 X2JDK3 O46040 A0A1A9XKB7 A0A1A9UU31 B5RJI3 B3MYS6 A0A1A9ZB74 B3P8Y6 A0A0A1XAS4 W8BTV1 A0A0Q5SS11 A0A1I8PSS9 A0A0K8V843 A0A1I8PSQ9 A0A0R3NXV9 A0A1I8PSV0 B4L5T8 A0A1B6C5C4 A0A1I8N6L9 B4NQ06 A0A1B0FK75 B4H372 A0A1Q3FS50 A0A1S3CTS0 A0A1J1IR78 A0A3Q0IJ68 B0XG67 T1PEZ7 A0A1I8N6M1 B5DKF2 A0A182FML8 A0A034W4S6 A0A182GQP9 A0A1W4VTU3 A0A0M3QZK5 A0A0Q5SY93 A0A1I8PSQ3 B4M7C7 P83501 W8BNI2 W5JT12 J9LMV0 A0A182PNS3 A0A0A1XGI4 B4L398 A0A0K8UI74 A0A3B0J935 A0A182MJM8 A0A182IZV8 A0A1I8N6M0 A0A1I8N6L8 A0A0A1WXV0 Q16ZX7 A0A1S4FI78 A0A1B0CEQ0 B4M3S5
A0A2H1WJV1 A0A2A4JQL2 D6WLG5 E2APG8 A0A1Y1MC08 A0A1Y1MDX9 A0A1Y1MF86 E9J2E4 A0A195B1H2 A0A195DJ28 A0A154P0F2 A0A3L8DDT9 A0A158NDG8 A0A195FI27 A0A151X0G6 A0A0N0U4Z4 A0A088AD93 A0A2A3ECJ9 A0A2P8ZCM0 A0A067QYV6 A0A1B6D0F2 A0A1B6E7R7 A0A0L7RA61 U4UGP7 A0A1B0CSC2 A0A1W4VT46 A0A1A9X4K0 A0A026WKD8 X2JDK3 O46040 A0A1A9XKB7 A0A1A9UU31 B5RJI3 B3MYS6 A0A1A9ZB74 B3P8Y6 A0A0A1XAS4 W8BTV1 A0A0Q5SS11 A0A1I8PSS9 A0A0K8V843 A0A1I8PSQ9 A0A0R3NXV9 A0A1I8PSV0 B4L5T8 A0A1B6C5C4 A0A1I8N6L9 B4NQ06 A0A1B0FK75 B4H372 A0A1Q3FS50 A0A1S3CTS0 A0A1J1IR78 A0A3Q0IJ68 B0XG67 T1PEZ7 A0A1I8N6M1 B5DKF2 A0A182FML8 A0A034W4S6 A0A182GQP9 A0A1W4VTU3 A0A0M3QZK5 A0A0Q5SY93 A0A1I8PSQ3 B4M7C7 P83501 W8BNI2 W5JT12 J9LMV0 A0A182PNS3 A0A0A1XGI4 B4L398 A0A0K8UI74 A0A3B0J935 A0A182MJM8 A0A182IZV8 A0A1I8N6M0 A0A1I8N6L8 A0A0A1WXV0 Q16ZX7 A0A1S4FI78 A0A1B0CEQ0 B4M3S5
PDB
3OXG
E-value=1.53824e-09,
Score=151
Ontologies
GO
Topology
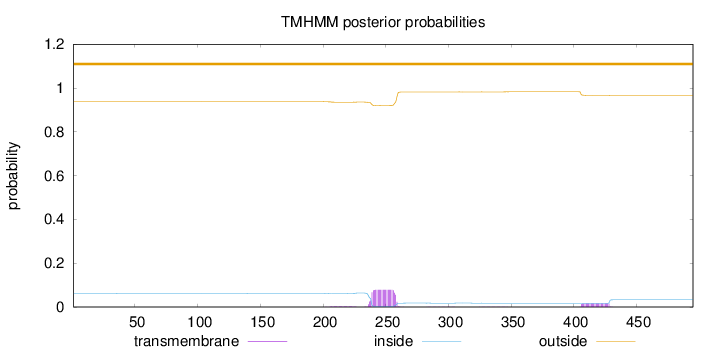
Length:
495
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.07262
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.06223
outside
1 - 495
Population Genetic Test Statistics
Pi
230.591031
Theta
166.075174
Tajima's D
0.993408
CLR
0.081938
CSRT
0.656417179141043
Interpretation
Uncertain
