Gene
KWMTBOMO02172 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003141
Annotation
PREDICTED:_probable_alpha-aspartyl_dipeptidase_[Amyelois_transitella]
Full name
Probable alpha-aspartyl dipeptidase
+ More
Fatty acyl-CoA reductase
Fatty acyl-CoA reductase
Alternative Name
Asp-specific dipeptidase
Location in the cell
PlasmaMembrane Reliability : 0.835
Sequence
CDS
ATGGCATCTCGTCGTCAAGCACTTTTGCTGTCATCATCCAACTGCCATGGATACAATCTTTTAGAATTTGCCAAAGCGGAAATCTCCAGTTTCCTCCAAAGGAACAATGTCAAGGATTTAATATTCATTCCTTATGCTCAAAATGACTTTGATGGCTATACTACTAAAATAAAGAATGTTATAGATCAATGGGGATTCACAGTCACTGGTCTGCACACATATCCTAAGCCAGAAACTGCTATTAGTGAATCTAAAGCTATCTTTATTGGGGGTGGAAATACTTTTTTACTTTTGAAAAGATTGTACAAAAATAACTTAGTCCAATTAATGAAAAGTAAAATTGATGATGGTAGTTTAATATACATTGGAAGCAGTGCGGGAACAAATGTAGCAACAAAAAGTATTCATACAACAAATGATATGCCAATAATTTATCCACCAAGTTTTGAAGCAATTGGCATAGTTCCATTCAACATTAATCCTCATTATATTGATAAAGTTGAAAATGAAGAACATAAAGGAGAGACAAGAGACCAAAGAATAGCTGAATATCTGGAGATGCAACATGCAGCTCCAGTGTTAGGACTGAAAGAGGGTGCAATATTGCATGTTGATGGTTCAAAACTTATCTTAAAAGGTGTAAATGGAGCAGTATTATTTAGTAGGAGTCAAAAGAAAAGTGAATATAATGTGGGAGCAGATTTATCTTTTTTATTAAATGAAATTTAA
Protein
MASRRQALLLSSSNCHGYNLLEFAKAEISSFLQRNNVKDLIFIPYAQNDFDGYTTKIKNVIDQWGFTVTGLHTYPKPETAISESKAIFIGGGNTFLLLKRLYKNNLVQLMKSKIDDGSLIYIGSSAGTNVATKSIHTTNDMPIIYPPSFEAIGIVPFNINPHYIDKVENEEHKGETRDQRIAEYLEMQHAAPVLGLKEGAILHVDGSKLILKGVNGAVLFSRSQKKSEYNVGADLSFLLNEI
Summary
Description
Hydrolyzes dipeptides containing N-terminal aspartate residues.
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalyzes the reduction of fatty acyl-CoA to fatty alcohols.
Catalytic Activity
Dipeptidase E catalyzes the hydrolysis of dipeptides Asp-|-Xaa. It does not act on peptides with N-terminal Glu, Asn or Gln, nor does it cleave isoaspartyl peptides.
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
a long-chain fatty acyl-CoA + 2 H(+) + 2 NADPH = a long-chain primary fatty alcohol + CoA + 2 NADP(+)
Similarity
Belongs to the peptidase S51 family.
Belongs to the fatty acyl-CoA reductase family.
Belongs to the fatty acyl-CoA reductase family.
Keywords
Complete proteome
Cytoplasm
Dipeptidase
Hydrolase
Protease
Reference proteome
Serine protease
Feature
chain Probable alpha-aspartyl dipeptidase
Uniprot
H9J0V4
A0A194PMX3
A0A194R433
S4NGQ4
A0A2H1WLQ0
A0A212ES62
+ More
A0A2A4JRF1 A0A2A4JQN5 A0A0L7LAS8 Q16FQ7 A0A182J9U8 A0A084VDN5 A0A023EL13 Q16ET3 A0A1Q3FDZ6 B0X281 A0A182H012 A0A182GW41 A0A023EN87 A0A182MZI1 A0A182Y7T3 A0A0T6AW81 A0A1Y1L5R1 A0A182QYY5 B4L622 A0A336LIB4 A0A182FMK7 A0A182KB05 A0A182PNR4 A0A182VLH4 A0A182SLH4 A0A0A1WNW6 A0A034WIC2 A0A182TJZ0 A0A2M4AQU8 A0A182XEG5 W5JQ60 A0A0K8V5B9 B4M727 B4JLY7 A0A0M4EM72 A0A1I8NA78 T1PFQ7 A0A182HGL3 N6TGL6 J3JX11 Q7QFI8 B4IGM1 A0A1I8Q5Q8 A0A1L8EFQ8 A0A1L8EG38 X2JER0 Q9VYH3 B3MS43 A0A1L8EG12 B4NUD4 A0A1W4XPS8 V5G8N2 B3NX71 A0A2L2YI25 A0A023EJT9 B4Q206 T1DPV3 W8ANQ4 A0A3B0KLB9 A0A1B6DYD8 Q29IB6 A0A1J1IXA5 A0A2P6KYE2 D6WJG1 A0A1S3KFN4 C3Z578 A0A182MKR7 A0A1B0A9T2 A0A182W4J7 A0A3B3XRY9 A0A087X443 A0A3B3UX92 A0A1B0CDZ3 A0A023EJA2 A0A1B0FL65 B4NEN4 A0A1W4VJH3 A0A1L8DTI3 A0A1B0B051 T1JMC9 U5EYH1 A0A2J7RDQ2 A0A1A9VC22 A0A3B5R6Q4 A0A0S7GK00 A0A146P492 A0A0K8R4E9 A0A067R009 A0A3P8TU73 A0A3B3ZIV7 B7PQV0 A0A1A9X544 H9G9B0 A0A0P4W6A0
A0A2A4JRF1 A0A2A4JQN5 A0A0L7LAS8 Q16FQ7 A0A182J9U8 A0A084VDN5 A0A023EL13 Q16ET3 A0A1Q3FDZ6 B0X281 A0A182H012 A0A182GW41 A0A023EN87 A0A182MZI1 A0A182Y7T3 A0A0T6AW81 A0A1Y1L5R1 A0A182QYY5 B4L622 A0A336LIB4 A0A182FMK7 A0A182KB05 A0A182PNR4 A0A182VLH4 A0A182SLH4 A0A0A1WNW6 A0A034WIC2 A0A182TJZ0 A0A2M4AQU8 A0A182XEG5 W5JQ60 A0A0K8V5B9 B4M727 B4JLY7 A0A0M4EM72 A0A1I8NA78 T1PFQ7 A0A182HGL3 N6TGL6 J3JX11 Q7QFI8 B4IGM1 A0A1I8Q5Q8 A0A1L8EFQ8 A0A1L8EG38 X2JER0 Q9VYH3 B3MS43 A0A1L8EG12 B4NUD4 A0A1W4XPS8 V5G8N2 B3NX71 A0A2L2YI25 A0A023EJT9 B4Q206 T1DPV3 W8ANQ4 A0A3B0KLB9 A0A1B6DYD8 Q29IB6 A0A1J1IXA5 A0A2P6KYE2 D6WJG1 A0A1S3KFN4 C3Z578 A0A182MKR7 A0A1B0A9T2 A0A182W4J7 A0A3B3XRY9 A0A087X443 A0A3B3UX92 A0A1B0CDZ3 A0A023EJA2 A0A1B0FL65 B4NEN4 A0A1W4VJH3 A0A1L8DTI3 A0A1B0B051 T1JMC9 U5EYH1 A0A2J7RDQ2 A0A1A9VC22 A0A3B5R6Q4 A0A0S7GK00 A0A146P492 A0A0K8R4E9 A0A067R009 A0A3P8TU73 A0A3B3ZIV7 B7PQV0 A0A1A9X544 H9G9B0 A0A0P4W6A0
EC Number
3.4.13.21
1.2.1.84
1.2.1.84
Pubmed
19121390
26354079
23622113
22118469
26227816
17510324
+ More
24438588 24945155 26483478 25244985 28004739 17994087 25830018 25348373 20920257 23761445 25315136 23537049 22516182 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 26561354 17550304 24495485 15632085 18362917 19820115 18563158 23542700 24845553 25463417 21881562
24438588 24945155 26483478 25244985 28004739 17994087 25830018 25348373 20920257 23761445 25315136 23537049 22516182 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 26561354 17550304 24495485 15632085 18362917 19820115 18563158 23542700 24845553 25463417 21881562
EMBL
BABH01010389
KQ459598
KPI94667.1
KQ460779
KPJ12284.1
GAIX01014644
+ More
JAA77916.1 ODYU01009145 SOQ53364.1 AGBW02012845 OWR44338.1 NWSH01000765 PCG74346.1 PCG74347.1 JTDY01001935 KOB72510.1 CH478397 EAT33071.1 ATLV01011631 KE524705 KFB36079.1 GAPW01003713 JAC09885.1 CH478591 EAT32743.1 EAT32744.1 EAT32745.1 GFDL01009259 JAV25786.1 DS232282 EDS39104.1 JXUM01100448 KQ564571 KXJ72080.1 JXUM01092375 KQ563983 KXJ72979.1 GAPW01003759 JAC09839.1 LJIG01022762 KRT78855.1 GEZM01063705 GEZM01063704 JAV69012.1 AXCN02001267 CH933812 EDW05818.1 UFQS01000015 UFQT01000015 SSW97386.1 SSX17772.1 GBXI01014092 JAD00200.1 GAKP01004524 GAKP01004523 JAC54429.1 GGFK01009828 MBW43149.1 ADMH02000428 ETN66542.1 GDHF01018187 JAI34127.1 CH940653 EDW62594.1 CH916371 EDV91748.1 CP012528 ALC48758.1 KA646738 AFP61367.1 APCN01005666 APGK01028295 KB740686 KB632224 ENN79539.1 ERL90217.1 BT127779 AEE62741.1 AAAB01008846 EAA06424.5 EGK96429.1 CH480836 EDW48971.1 GFDG01001258 JAV17541.1 GFDG01001256 JAV17543.1 AE014298 AHN59653.1 AY118855 CH902622 EDV34598.1 GFDG01001260 JAV17539.1 CH983901 EDX16581.1 GALX01002001 JAB66465.1 CH954180 EDV47243.1 IAAA01020362 IAAA01020363 LAA07782.1 GAPW01004252 JAC09346.1 CM000162 EDX01527.1 GAMD01001630 JAA99960.1 GAMC01020462 JAB86093.1 OUUW01000021 SPP89410.1 GEDC01029569 GEDC01012027 GEDC01011685 GEDC01006617 JAS07729.1 JAS25271.1 JAS25613.1 JAS30681.1 CH379063 EAL32737.3 CVRI01000063 CRL04338.1 MWRG01003648 PRD31341.1 KQ971342 EFA03678.1 GG666582 EEN52435.1 AXCM01011571 AYCK01019428 AJWK01008435 AJWK01008436 AJWK01008437 GAPW01004561 JAC09037.1 CCAG010017382 CH964239 EDW82203.1 GFDF01004321 JAV09763.1 JXJN01006562 JH431934 GANO01002004 JAB57867.1 NEVH01005285 PNF38963.1 GBYX01452042 JAO29468.1 GCES01147563 JAQ38759.1 GADI01008464 JAA65344.1 KK852840 KDR15187.1 ABJB010092262 DS767852 EEC08972.1 AAWZ02015295 GDRN01097444 JAI59114.1
JAA77916.1 ODYU01009145 SOQ53364.1 AGBW02012845 OWR44338.1 NWSH01000765 PCG74346.1 PCG74347.1 JTDY01001935 KOB72510.1 CH478397 EAT33071.1 ATLV01011631 KE524705 KFB36079.1 GAPW01003713 JAC09885.1 CH478591 EAT32743.1 EAT32744.1 EAT32745.1 GFDL01009259 JAV25786.1 DS232282 EDS39104.1 JXUM01100448 KQ564571 KXJ72080.1 JXUM01092375 KQ563983 KXJ72979.1 GAPW01003759 JAC09839.1 LJIG01022762 KRT78855.1 GEZM01063705 GEZM01063704 JAV69012.1 AXCN02001267 CH933812 EDW05818.1 UFQS01000015 UFQT01000015 SSW97386.1 SSX17772.1 GBXI01014092 JAD00200.1 GAKP01004524 GAKP01004523 JAC54429.1 GGFK01009828 MBW43149.1 ADMH02000428 ETN66542.1 GDHF01018187 JAI34127.1 CH940653 EDW62594.1 CH916371 EDV91748.1 CP012528 ALC48758.1 KA646738 AFP61367.1 APCN01005666 APGK01028295 KB740686 KB632224 ENN79539.1 ERL90217.1 BT127779 AEE62741.1 AAAB01008846 EAA06424.5 EGK96429.1 CH480836 EDW48971.1 GFDG01001258 JAV17541.1 GFDG01001256 JAV17543.1 AE014298 AHN59653.1 AY118855 CH902622 EDV34598.1 GFDG01001260 JAV17539.1 CH983901 EDX16581.1 GALX01002001 JAB66465.1 CH954180 EDV47243.1 IAAA01020362 IAAA01020363 LAA07782.1 GAPW01004252 JAC09346.1 CM000162 EDX01527.1 GAMD01001630 JAA99960.1 GAMC01020462 JAB86093.1 OUUW01000021 SPP89410.1 GEDC01029569 GEDC01012027 GEDC01011685 GEDC01006617 JAS07729.1 JAS25271.1 JAS25613.1 JAS30681.1 CH379063 EAL32737.3 CVRI01000063 CRL04338.1 MWRG01003648 PRD31341.1 KQ971342 EFA03678.1 GG666582 EEN52435.1 AXCM01011571 AYCK01019428 AJWK01008435 AJWK01008436 AJWK01008437 GAPW01004561 JAC09037.1 CCAG010017382 CH964239 EDW82203.1 GFDF01004321 JAV09763.1 JXJN01006562 JH431934 GANO01002004 JAB57867.1 NEVH01005285 PNF38963.1 GBYX01452042 JAO29468.1 GCES01147563 JAQ38759.1 GADI01008464 JAA65344.1 KK852840 KDR15187.1 ABJB010092262 DS767852 EEC08972.1 AAWZ02015295 GDRN01097444 JAI59114.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000037510
+ More
UP000008820 UP000075880 UP000030765 UP000002320 UP000069940 UP000249989 UP000075884 UP000076408 UP000075886 UP000009192 UP000069272 UP000075881 UP000075885 UP000075903 UP000075901 UP000075902 UP000076407 UP000000673 UP000008792 UP000001070 UP000092553 UP000095301 UP000075840 UP000019118 UP000030742 UP000007062 UP000001292 UP000095300 UP000000803 UP000007801 UP000000304 UP000192223 UP000008711 UP000002282 UP000268350 UP000001819 UP000183832 UP000007266 UP000085678 UP000001554 UP000075883 UP000092445 UP000075920 UP000261480 UP000028760 UP000261500 UP000092461 UP000092444 UP000007798 UP000192221 UP000092460 UP000235965 UP000078200 UP000002852 UP000027135 UP000265080 UP000261520 UP000001555 UP000091820 UP000001646
UP000008820 UP000075880 UP000030765 UP000002320 UP000069940 UP000249989 UP000075884 UP000076408 UP000075886 UP000009192 UP000069272 UP000075881 UP000075885 UP000075903 UP000075901 UP000075902 UP000076407 UP000000673 UP000008792 UP000001070 UP000092553 UP000095301 UP000075840 UP000019118 UP000030742 UP000007062 UP000001292 UP000095300 UP000000803 UP000007801 UP000000304 UP000192223 UP000008711 UP000002282 UP000268350 UP000001819 UP000183832 UP000007266 UP000085678 UP000001554 UP000075883 UP000092445 UP000075920 UP000261480 UP000028760 UP000261500 UP000092461 UP000092444 UP000007798 UP000192221 UP000092460 UP000235965 UP000078200 UP000002852 UP000027135 UP000265080 UP000261520 UP000001555 UP000091820 UP000001646
Interpro
Gene 3D
ProteinModelPortal
H9J0V4
A0A194PMX3
A0A194R433
S4NGQ4
A0A2H1WLQ0
A0A212ES62
+ More
A0A2A4JRF1 A0A2A4JQN5 A0A0L7LAS8 Q16FQ7 A0A182J9U8 A0A084VDN5 A0A023EL13 Q16ET3 A0A1Q3FDZ6 B0X281 A0A182H012 A0A182GW41 A0A023EN87 A0A182MZI1 A0A182Y7T3 A0A0T6AW81 A0A1Y1L5R1 A0A182QYY5 B4L622 A0A336LIB4 A0A182FMK7 A0A182KB05 A0A182PNR4 A0A182VLH4 A0A182SLH4 A0A0A1WNW6 A0A034WIC2 A0A182TJZ0 A0A2M4AQU8 A0A182XEG5 W5JQ60 A0A0K8V5B9 B4M727 B4JLY7 A0A0M4EM72 A0A1I8NA78 T1PFQ7 A0A182HGL3 N6TGL6 J3JX11 Q7QFI8 B4IGM1 A0A1I8Q5Q8 A0A1L8EFQ8 A0A1L8EG38 X2JER0 Q9VYH3 B3MS43 A0A1L8EG12 B4NUD4 A0A1W4XPS8 V5G8N2 B3NX71 A0A2L2YI25 A0A023EJT9 B4Q206 T1DPV3 W8ANQ4 A0A3B0KLB9 A0A1B6DYD8 Q29IB6 A0A1J1IXA5 A0A2P6KYE2 D6WJG1 A0A1S3KFN4 C3Z578 A0A182MKR7 A0A1B0A9T2 A0A182W4J7 A0A3B3XRY9 A0A087X443 A0A3B3UX92 A0A1B0CDZ3 A0A023EJA2 A0A1B0FL65 B4NEN4 A0A1W4VJH3 A0A1L8DTI3 A0A1B0B051 T1JMC9 U5EYH1 A0A2J7RDQ2 A0A1A9VC22 A0A3B5R6Q4 A0A0S7GK00 A0A146P492 A0A0K8R4E9 A0A067R009 A0A3P8TU73 A0A3B3ZIV7 B7PQV0 A0A1A9X544 H9G9B0 A0A0P4W6A0
A0A2A4JRF1 A0A2A4JQN5 A0A0L7LAS8 Q16FQ7 A0A182J9U8 A0A084VDN5 A0A023EL13 Q16ET3 A0A1Q3FDZ6 B0X281 A0A182H012 A0A182GW41 A0A023EN87 A0A182MZI1 A0A182Y7T3 A0A0T6AW81 A0A1Y1L5R1 A0A182QYY5 B4L622 A0A336LIB4 A0A182FMK7 A0A182KB05 A0A182PNR4 A0A182VLH4 A0A182SLH4 A0A0A1WNW6 A0A034WIC2 A0A182TJZ0 A0A2M4AQU8 A0A182XEG5 W5JQ60 A0A0K8V5B9 B4M727 B4JLY7 A0A0M4EM72 A0A1I8NA78 T1PFQ7 A0A182HGL3 N6TGL6 J3JX11 Q7QFI8 B4IGM1 A0A1I8Q5Q8 A0A1L8EFQ8 A0A1L8EG38 X2JER0 Q9VYH3 B3MS43 A0A1L8EG12 B4NUD4 A0A1W4XPS8 V5G8N2 B3NX71 A0A2L2YI25 A0A023EJT9 B4Q206 T1DPV3 W8ANQ4 A0A3B0KLB9 A0A1B6DYD8 Q29IB6 A0A1J1IXA5 A0A2P6KYE2 D6WJG1 A0A1S3KFN4 C3Z578 A0A182MKR7 A0A1B0A9T2 A0A182W4J7 A0A3B3XRY9 A0A087X443 A0A3B3UX92 A0A1B0CDZ3 A0A023EJA2 A0A1B0FL65 B4NEN4 A0A1W4VJH3 A0A1L8DTI3 A0A1B0B051 T1JMC9 U5EYH1 A0A2J7RDQ2 A0A1A9VC22 A0A3B5R6Q4 A0A0S7GK00 A0A146P492 A0A0K8R4E9 A0A067R009 A0A3P8TU73 A0A3B3ZIV7 B7PQV0 A0A1A9X544 H9G9B0 A0A0P4W6A0
PDB
1FYE
E-value=7.49313e-35,
Score=366
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
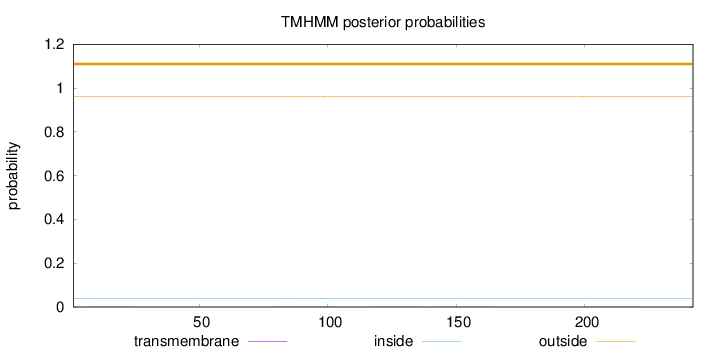
Length:
242
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03423
Exp number, first 60 AAs:
0.00334
Total prob of N-in:
0.03968
outside
1 - 242
Population Genetic Test Statistics
Pi
180.847596
Theta
211.18685
Tajima's D
-0.352541
CLR
24.452334
CSRT
0.278986050697465
Interpretation
Uncertain
