Gene
KWMTBOMO02171
Pre Gene Modal
BGIBMGA003023
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase_G-like_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.238
Sequence
CDS
ATGTCAAAACAAGGAAAAAGAAAAATATCTGTTGAATTGCCGAAAACGAAAGATCTCTCTATTAGACCTAGTGTGTTTGATCGGCTCGGTACTAAGAAATCATCAAGTGCTAAAAAAACAACTGAACATTGTAGACAGTGGGCCCAACACGGCAGCTGTGCTTACGGCAAAAGTTGTAAATTTGCTTCTACGCATACTCTTATCAGTCCTTCTAAACAACGCGCAGCTCATAAAGACAATGATCAGAAACGTTTAGTGAAGGATGATGGGAAGACTAGATTGCATTCTACTGTTATAGTGCGGTCAGGTCAAAGCCCCGAAGGAGAAATCGATAACTGGGACCAAAATGACTTAGAGTATGCAGACACAGATGTCCTCGAAAAACGAAGACAGCAGTTGCAGCGTGAACTAGAATTACAGATGAAAATGGATTCAAACAAAGACATGCGAAAAGATAAAAAAAAAGTCGTGTCCAGTACGTCTTCATCTAGAAGCTCTAGTTCTTCGAGTGTTTCGTCGTCTTCATCAGACGGCAGCACATCGTCATCTTCATCTGGAAGGGAAACTAGAAGGAAAACTAAAAAAGGAAAACTTAAACGAAATTCCAGTTCTTCTTCAGATGGAAACATACCAACAAAAAAGAAATACAATGTGAAGAGTGAACTGTCCAAAAAAGATAAAAGTGATGTTAAAAAAAATATTACAAAAAAAGATGATAGATTAATAAAAAAGAGTGATAGTTCCAAAAAGAAATCAACTTCTAAAACTCCCATAGGAAAAAAGTCACTATCTCCAGGTAAAAAGTCTGGTGGTACTCCACCCATAACATCCAAGACACTTATTAAAAGTTCGTCTAAACACAGTAAATCACCCTTAAGAAGAGATAGGAATAGAAGTGTGTCTCCACATTTTAGGGATAAAGAAAAAGAACGAGAACGTAATAAAGAGAAAGAAAGAGAAAAAGATAGAGATAAGGATAGAGAAAAAGAAAGAGACAGAAACAGAGCACGTAGTAGAACTCCACGAAAAGGTCGTTCGAGATCTCCTAGACAAAATGCAAAGGATAATAGAAAAAGGGAGAGTCCTGAGAGAGGCAGGCCTGTGTCACCCAAAAAGCAATCCCGCAGAGATGGCAGTCCTGATAGACATCGAAAACGGTCTACTAGTCGTGATAGGGAGAGAGGTCGTGATCATAAAGATAGATCCTTAGAGAGAAGAAAAGAAAAATATGATGATAAAGACAGGCACGATCGTAAAGATAGGGGTCGCGAGCGAAAAGAGGTTAAACGGGATGATAGTAAAAGAAGGGGTCGTGAACGAGGTCTTGGAGGAAGAAGTACAGGTGATAAAGGTTTAGAAAAACCAAATAAACCGATGGAGAGACTTCTTCCAAGACCTGAAGAACGTCTAGCTGCACTAGCTGCCATTTCAAATAGAACTCTGGAATCAGATAAAAACTCGAATACATCAAAAGATAGACAAGATACACATTCTGAACGCGGAGGTAGGAAGCCAGAACGACTTGAGAGAGAGCGGTCTAGTAAACGAGATCGCTTGGGCAGCAATGACAGAGAACAGGGTGAATACAGTATGGGTATTGACAGACAATATGATGATCATGGCATTGACCATTATGATAGAGTTAACGATCGTTATGATGGCGTTACTAGAGATGACGATCGGTCACCTGGCTATGGTGCACAAGGGCGAGAAAGACACTATGAAGCGCCATATGAAATTGCTGGTCCGCCGAGAGGATTTCCTGAAGAAGATGATAGAATCTATAATGAACATATGGGAGATCACAGAGATGTTGAAATGTGTTATAATGAAAGACGACCACATCGAGATCGTTCTTGGGAAGGACGTTCAGGCTCTCTTGATCGTGAACGTGCATACATGCATCGAAAAGATTGGGACAATGAAGATTATCATGCCAGCGGCGACTGGGGTCGAGATAGACACTGGCCTATTCATGATTCTCAGATGAGAGAATGGGGTGATAAAGATCCAGAAGATAATTGGCACCATAGAGGTCATCCTGGTCAACATCGTGGTCGAATACATGATGAGTATGGTCGAGGCATGAGATTAGATGTCGATCGCGGTGACAAACGTCGAAATGTTCGAAATGAACCTGATCAAAGTTCGAAAGATACCGGTCATCCTCAAACTTCTACAGCGTCTACTCGTCAAGAAACGGAAATTGAAGAAAAACCAATACCCGATGATCTTAGTGAAATCAGTGATGATCCTGATGATATATTAAATAGAGAAGATATGATGGATCAAAATAATGAAGAAGAAACTCAAAACCTGTCTGAAGAAGTGGCAATAAAGCCTGAAAGTATAGAAAAAGTGTCCGAGCAGGATGCAAGTGCTGTTGGAGACGAAAAAGAAATACAAGATAACAAAGATGATGAAGATGTTGCAAACTTAGACTTTGAGGAAATATCTGATGGAGAACTGGAAGAGGAAAAAACAAGAGCAGGATTAGGTGACGCACTTGGCGTCGATTGGGCAAGCTTAGTAGCGGATGTGCGGCGTCGGGAACAAACAACGCCTACAAGCGGAGCGCGTGAGCGCTGGCGACCAGAGAGGGTACTTGCACGAGTAGGTCTTTCTGTACATTTGGCTGGTAAAGATTTGGTGAGCGATATATTAAAAAACAACAAAGAATTACTCCAAGCTGAGAAGACTGCTCAAGATCAGAAATGCGAAAAATCTGAAAACTTGGTGCAAGAGAATGGTTGTCATGATATCCAAACAAGAAAGCTTGAAGATATTTCAGATATTCAGATTTTACATCCAGTAGCTTCTATACAGGTGGCTATGGAAAAGAGGAAACGTCAACGTGCTGTTTTGTTTGGATCAGGATGCGAGATTACCAGAGGTCTGAGTGCAAGGCGCGATTTGGCGTTACGCCGTTATTTATGTCACTTGCCAGCAACAGAACGAACTGGACAGTCTCGTGTTACACCACAGCCTTCACTGTTTGAGGCTGCTAAAGCTCTCTTGATGCGGGCTCCTGTCTCTGGTTAA
Protein
MSKQGKRKISVELPKTKDLSIRPSVFDRLGTKKSSSAKKTTEHCRQWAQHGSCAYGKSCKFASTHTLISPSKQRAAHKDNDQKRLVKDDGKTRLHSTVIVRSGQSPEGEIDNWDQNDLEYADTDVLEKRRQQLQRELELQMKMDSNKDMRKDKKKVVSSTSSSRSSSSSSVSSSSSDGSTSSSSSGRETRRKTKKGKLKRNSSSSSDGNIPTKKKYNVKSELSKKDKSDVKKNITKKDDRLIKKSDSSKKKSTSKTPIGKKSLSPGKKSGGTPPITSKTLIKSSSKHSKSPLRRDRNRSVSPHFRDKEKERERNKEKEREKDRDKDREKERDRNRARSRTPRKGRSRSPRQNAKDNRKRESPERGRPVSPKKQSRRDGSPDRHRKRSTSRDRERGRDHKDRSLERRKEKYDDKDRHDRKDRGRERKEVKRDDSKRRGRERGLGGRSTGDKGLEKPNKPMERLLPRPEERLAALAAISNRTLESDKNSNTSKDRQDTHSERGGRKPERLERERSSKRDRLGSNDREQGEYSMGIDRQYDDHGIDHYDRVNDRYDGVTRDDDRSPGYGAQGRERHYEAPYEIAGPPRGFPEEDDRIYNEHMGDHRDVEMCYNERRPHRDRSWEGRSGSLDRERAYMHRKDWDNEDYHASGDWGRDRHWPIHDSQMREWGDKDPEDNWHHRGHPGQHRGRIHDEYGRGMRLDVDRGDKRRNVRNEPDQSSKDTGHPQTSTASTRQETEIEEKPIPDDLSEISDDPDDILNREDMMDQNNEEETQNLSEEVAIKPESIEKVSEQDASAVGDEKEIQDNKDDEDVANLDFEEISDGELEEEKTRAGLGDALGVDWASLVADVRRREQTTPTSGARERWRPERVLARVGLSVHLAGKDLVSDILKNNKELLQAEKTAQDQKCEKSENLVQENGCHDIQTRKLEDISDIQILHPVASIQVAMEKRKRQRAVLFGSGCEITRGLSARRDLALRRYLCHLPATERTGQSRVTPQPSLFEAAKALLMRAPVSG
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00642 zf-CCCH
ProteinModelPortal
Ontologies
GO
PANTHER
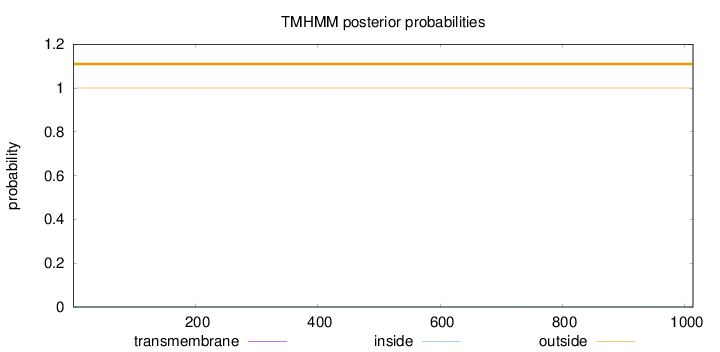
Topology
Length:
1013
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00105
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.00003
outside
1 - 1013
Population Genetic Test Statistics
Pi
168.943517
Theta
172.196782
Tajima's D
-1.234163
CLR
0.616397
CSRT
0.0984950752462377
Interpretation
Uncertain
