Gene
KWMTBOMO02167 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003021
Annotation
Protein_unzipped_[Papilio_xuthus]
Full name
Protein unzipped
Alternative Name
Protein zipper
Location in the cell
PlasmaMembrane Reliability : 2.332
Sequence
CDS
ATGAAAAACAGAAGTCGTCAAAGCGGTTGGTGGATATTGCTAGTAACAATGGCGCTGATTGCACCGGCTGCATCTGAGGCTGGAGTTAACCTTTTCAATTCAAGATTGAAACAAATCACTACTTCAAGCACTCTGAAATGGAAGAATTTCAGAAAAGAATTCATGAATAATTACGTTGTTGGAGCAGAATCTGAAGAAGGTCCCATATATCTCTGCAGAGCTCGACATGAAGGAGATATGATACTTGGACAATTGAAGCCAGATTATAGTTCTTGTGCTGTTTCTGGGAGCAAGAGCTACAGCATATTCGAAGTACTTGAAAATATAGAAAACGCATCTTTAATTGATTGGATCCCATGGTTTAAATTTAATGCAAGACCTCACGGAGCTGTAGCAAGTTCTTTTAGTGATTCTGTGTTTATTGGGAGAATGAAAGCTTCAAGCGGTTATTCGCATTACATAGGAAGAATCGTTTATGAAAGTGCTATTGGACGACTTATAACGTTTGATGATAAAAACAACGACTTAATTGAAAACAGCGGTGACATTTTAGTAGAAATAGAACCAGTTAGTTATAAATTAGAAGATGTTCAATTAGATTTAAAAACTGAACATATACGTCAAAACGATCCTGAGGTATTCGAAGTACGAACTCTAAGAAACGATGAAGAACTTGGCGCTACAGTTACCACAGAAATTGAATACGCTTATAACTACACTGTTTCTTGGGGCCACGGGCACGGCATAGCTATTGGTTTGAACACTACCATTGAAATGGCAGATGGTGATGTTTTACCGCCAATCCAATGGGCTAATCCGTTCACAGAAAGGCGAACCAGGTTGCATAAGCTTGAAAAATATTTAGAACCAGGGACCGCTTGCAATGTAACATTCAGTGGCAACCGTACGGCTAGAGATGTCGAATATACCGCTAAGCTGATCACCTTTTACAATGGAAAAGACCCACGCTCTCGTCAAATGCGTGGACAACGTATAGAAAATACATTAGAAGTTGAGGCAGTTTATGGTGATATATACTTTCTTGGCAACAGCAGCTTAGTACCTACTACTACAACTACAACCACAACTACCACAACCACCACTACCACAACACCTGCTCCAACCGTTCCTCCAGCACCTGAGGAAAACGATATCGCAAGTCCAATGAAAAAACCAAACACACCGGTGGAAGTAGATAGCAACGATATGATGGGTGATAGCGGAGAAGGAATGATTAAAACACCTCCAGTAAAAGATGATAATATCAACCGTTCCGTTGTGGGTGGCCTTGGCAGTACACCTAATTCGGGTAACACAATGCGATTTTTTTACGTTACATTAGTACTCCCAATATTTTCTTTTTTCCATTAA
Protein
MKNRSRQSGWWILLVTMALIAPAASEAGVNLFNSRLKQITTSSTLKWKNFRKEFMNNYVVGAESEEGPIYLCRARHEGDMILGQLKPDYSSCAVSGSKSYSIFEVLENIENASLIDWIPWFKFNARPHGAVASSFSDSVFIGRMKASSGYSHYIGRIVYESAIGRLITFDDKNNDLIENSGDILVEIEPVSYKLEDVQLDLKTEHIRQNDPEVFEVRTLRNDEELGATVTTEIEYAYNYTVSWGHGHGIAIGLNTTIEMADGDVLPPIQWANPFTERRTRLHKLEKYLEPGTACNVTFSGNRTARDVEYTAKLITFYNGKDPRSRQMRGQRIENTLEVEAVYGDIYFLGNSSLVPTTTTTTTTTTTTTTTTPAPTVPPAPEENDIASPMKKPNTPVEVDSNDMMGDSGEGMIKTPPVKDDNINRSVVGGLGSTPNSGNTMRFFYVTLVLPIFSFFH
Summary
Keywords
Complete proteome
Developmental protein
Differentiation
Glycoprotein
Membrane
Neurogenesis
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Protein unzipped
Uniprot
H9J0I4
A0A2A4JSG7
A0A2H1V3V6
A0A194PMX8
A0A0N1PHN4
A0A0L7LU66
+ More
A0A212ES78 A0A1E1W9M6 A0A067QXL4 A0A0M8ZRL2 A0A154PJB8 A0A2J7QYZ9 A0A0L7R8N2 F4WZX8 A0A2P8YV38 A0A158NGP6 V9IJU4 A0A151IFH0 T1HED6 A0A2A3E3U6 A0A195FFA6 Q4PKR8 A0A088A6G6 A0A1B6E9E8 A0A151XAE5 E2BD97 A0A1S3D5F2 E9ILB7 D6WN71 A0A1Y1M100 E1ZV65 A0A0T6BG16 N6UFY3 A0A2R7VV21 A0A1W4XIY9 A0A1W4XJX4 B4IHC5 B4QD58 B3NR96 A0A1W4V169 B4PBZ2 A0A0B4LGL6 P10379 A0A1L8DXA3 A0A0K8WAB7 B3MCD4 A0A0A1XP79 A0A1B0CIB2 A0A034VPI8 A0A1A9WV60 A0A3B0JJ97 B4MPM2 A0A3B0J1G1 B4GI31 Q28Z91 B4LP61 W8AJI0 A0A1A9Y731 A0A1I8MTT6 T1PFY7 A0A1B0BGM9 A0A1I8NMY9 A0A1B0FPQ9 A0A1A9Z5C1 B4LA33 A0A1A9VVQ2 A0A084VIF9 A0A0M4EF47 A0A336MUV1 A0A182QP96 A0A182N2H9 A0A336M9W1 A0A0N8DIT6 A0A0P6JJ41 A0A164X310 A0A0P5LK76 A0A182R715 A0A182IJB7 A0A0P6GED9 A0A023ET71 A0A182GZ10 A0A182TRJ3 Q17N01 A0A1Y9GKG9 Q7PZ21
A0A212ES78 A0A1E1W9M6 A0A067QXL4 A0A0M8ZRL2 A0A154PJB8 A0A2J7QYZ9 A0A0L7R8N2 F4WZX8 A0A2P8YV38 A0A158NGP6 V9IJU4 A0A151IFH0 T1HED6 A0A2A3E3U6 A0A195FFA6 Q4PKR8 A0A088A6G6 A0A1B6E9E8 A0A151XAE5 E2BD97 A0A1S3D5F2 E9ILB7 D6WN71 A0A1Y1M100 E1ZV65 A0A0T6BG16 N6UFY3 A0A2R7VV21 A0A1W4XIY9 A0A1W4XJX4 B4IHC5 B4QD58 B3NR96 A0A1W4V169 B4PBZ2 A0A0B4LGL6 P10379 A0A1L8DXA3 A0A0K8WAB7 B3MCD4 A0A0A1XP79 A0A1B0CIB2 A0A034VPI8 A0A1A9WV60 A0A3B0JJ97 B4MPM2 A0A3B0J1G1 B4GI31 Q28Z91 B4LP61 W8AJI0 A0A1A9Y731 A0A1I8MTT6 T1PFY7 A0A1B0BGM9 A0A1I8NMY9 A0A1B0FPQ9 A0A1A9Z5C1 B4LA33 A0A1A9VVQ2 A0A084VIF9 A0A0M4EF47 A0A336MUV1 A0A182QP96 A0A182N2H9 A0A336M9W1 A0A0N8DIT6 A0A0P6JJ41 A0A164X310 A0A0P5LK76 A0A182R715 A0A182IJB7 A0A0P6GED9 A0A023ET71 A0A182GZ10 A0A182TRJ3 Q17N01 A0A1Y9GKG9 Q7PZ21
Pubmed
19121390
26354079
26227816
22118469
24845553
21719571
+ More
29403074 21347285 20798317 21282665 18362917 19820115 28004739 23537049 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3402433 12537569 17893096 19349973 18057021 25830018 25348373 15632085 23185243 24495485 25315136 24438588 24945155 26483478 17510324 12364791 14747013 17210077
29403074 21347285 20798317 21282665 18362917 19820115 28004739 23537049 17994087 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 3402433 12537569 17893096 19349973 18057021 25830018 25348373 15632085 23185243 24495485 25315136 24438588 24945155 26483478 17510324 12364791 14747013 17210077
EMBL
BABH01010395
NWSH01000765
PCG74342.1
ODYU01000372
SOQ35042.1
KQ459598
+ More
KPI94672.1 KQ460709 KPJ12737.1 JTDY01000079 KOB78992.1 AGBW02012845 OWR44343.1 GDQN01007344 JAT83710.1 KK852840 KDR15190.1 KQ435922 KOX68546.1 KQ434936 KZC11917.1 NEVH01009081 PNF33808.1 KQ414632 KOC67199.1 GL888480 EGI60268.1 PYGN01000343 PSN48106.1 ADTU01015164 JR049312 AEY60927.1 KQ977793 KYM99646.1 ACPB03010241 KZ288405 PBC26164.1 KQ981625 KYN39068.1 DQ071552 AAY82248.1 GEDC01002747 JAS34551.1 KQ982339 KYQ57336.1 GL447585 EFN86325.1 GL764074 EFZ18643.1 KQ971342 EFA03053.1 GEZM01046052 JAV77556.1 GL434441 EFN74924.1 LJIG01000831 KRT86109.1 APGK01028301 KB740686 KB631083 KB632224 ENN79541.1 ERL84084.1 ERL90219.1 KK854087 PTY11008.1 CH480838 EDW49301.1 CM000362 CM002911 EDX08654.1 KMY96513.1 CH954179 EDV57115.2 CM000158 EDW92646.2 AE013599 AHN56670.1 X07450 AY052139 GFDF01002997 JAV11087.1 GDHF01018426 GDHF01014036 GDHF01012455 GDHF01004554 JAI33888.1 JAI38278.1 JAI39859.1 JAI47760.1 CH902619 EDV37258.1 KPU76662.1 GBXI01010492 GBXI01005542 GBXI01002385 GBXI01001545 JAD03800.1 JAD08750.1 JAD11907.1 JAD12747.1 AJWK01013172 GAKP01014560 GAKP01014559 GAKP01014558 GAKP01014557 GAKP01014556 GAKP01014555 GAKP01014554 JAC44394.1 OUUW01000001 SPP72671.1 CH963849 EDW74061.1 SPP72672.1 CH479183 EDW36151.1 CM000071 EAL25723.3 KRT02483.1 CH940648 EDW60170.2 KRF79286.1 GAMC01017850 JAB88705.1 KA647671 AFP62300.1 JXJN01013911 CCAG010023635 CH933882 EDW05468.2 ATLV01013356 KE524854 KFB37753.1 CP012524 ALC42815.1 UFQT01001856 SSX32027.1 AXCN02001074 AXCN02001075 UFQS01000766 UFQT01000766 SSX06720.1 SSX27066.1 GDIP01029559 JAM74156.1 GDIQ01017438 JAN77299.1 LRGB01001021 KZS13823.1 GDIQ01168424 JAK83301.1 GDIQ01034322 JAN60415.1 GAPW01001083 JAC12515.1 JXUM01098494 JXUM01098495 KQ564424 KXJ72309.1 CH477203 EAT48001.1 APCN01002368 AAAB01008986 EAA00798.4
KPI94672.1 KQ460709 KPJ12737.1 JTDY01000079 KOB78992.1 AGBW02012845 OWR44343.1 GDQN01007344 JAT83710.1 KK852840 KDR15190.1 KQ435922 KOX68546.1 KQ434936 KZC11917.1 NEVH01009081 PNF33808.1 KQ414632 KOC67199.1 GL888480 EGI60268.1 PYGN01000343 PSN48106.1 ADTU01015164 JR049312 AEY60927.1 KQ977793 KYM99646.1 ACPB03010241 KZ288405 PBC26164.1 KQ981625 KYN39068.1 DQ071552 AAY82248.1 GEDC01002747 JAS34551.1 KQ982339 KYQ57336.1 GL447585 EFN86325.1 GL764074 EFZ18643.1 KQ971342 EFA03053.1 GEZM01046052 JAV77556.1 GL434441 EFN74924.1 LJIG01000831 KRT86109.1 APGK01028301 KB740686 KB631083 KB632224 ENN79541.1 ERL84084.1 ERL90219.1 KK854087 PTY11008.1 CH480838 EDW49301.1 CM000362 CM002911 EDX08654.1 KMY96513.1 CH954179 EDV57115.2 CM000158 EDW92646.2 AE013599 AHN56670.1 X07450 AY052139 GFDF01002997 JAV11087.1 GDHF01018426 GDHF01014036 GDHF01012455 GDHF01004554 JAI33888.1 JAI38278.1 JAI39859.1 JAI47760.1 CH902619 EDV37258.1 KPU76662.1 GBXI01010492 GBXI01005542 GBXI01002385 GBXI01001545 JAD03800.1 JAD08750.1 JAD11907.1 JAD12747.1 AJWK01013172 GAKP01014560 GAKP01014559 GAKP01014558 GAKP01014557 GAKP01014556 GAKP01014555 GAKP01014554 JAC44394.1 OUUW01000001 SPP72671.1 CH963849 EDW74061.1 SPP72672.1 CH479183 EDW36151.1 CM000071 EAL25723.3 KRT02483.1 CH940648 EDW60170.2 KRF79286.1 GAMC01017850 JAB88705.1 KA647671 AFP62300.1 JXJN01013911 CCAG010023635 CH933882 EDW05468.2 ATLV01013356 KE524854 KFB37753.1 CP012524 ALC42815.1 UFQT01001856 SSX32027.1 AXCN02001074 AXCN02001075 UFQS01000766 UFQT01000766 SSX06720.1 SSX27066.1 GDIP01029559 JAM74156.1 GDIQ01017438 JAN77299.1 LRGB01001021 KZS13823.1 GDIQ01168424 JAK83301.1 GDIQ01034322 JAN60415.1 GAPW01001083 JAC12515.1 JXUM01098494 JXUM01098495 KQ564424 KXJ72309.1 CH477203 EAT48001.1 APCN01002368 AAAB01008986 EAA00798.4
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000027135 UP000053105 UP000076502 UP000235965 UP000053825 UP000007755 UP000245037 UP000005205 UP000078542 UP000015103 UP000242457 UP000078541 UP000005203 UP000075809 UP000008237 UP000079169 UP000007266 UP000000311 UP000019118 UP000030742 UP000192223 UP000001292 UP000000304 UP000008711 UP000192221 UP000002282 UP000000803 UP000007801 UP000092461 UP000091820 UP000268350 UP000007798 UP000008744 UP000001819 UP000008792 UP000092443 UP000095301 UP000092460 UP000095300 UP000092444 UP000092445 UP000009192 UP000078200 UP000030765 UP000092553 UP000075886 UP000075884 UP000076858 UP000075900 UP000075880 UP000069940 UP000249989 UP000075902 UP000008820 UP000075840 UP000007062
UP000027135 UP000053105 UP000076502 UP000235965 UP000053825 UP000007755 UP000245037 UP000005205 UP000078542 UP000015103 UP000242457 UP000078541 UP000005203 UP000075809 UP000008237 UP000079169 UP000007266 UP000000311 UP000019118 UP000030742 UP000192223 UP000001292 UP000000304 UP000008711 UP000192221 UP000002282 UP000000803 UP000007801 UP000092461 UP000091820 UP000268350 UP000007798 UP000008744 UP000001819 UP000008792 UP000092443 UP000095301 UP000092460 UP000095300 UP000092444 UP000092445 UP000009192 UP000078200 UP000030765 UP000092553 UP000075886 UP000075884 UP000076858 UP000075900 UP000075880 UP000069940 UP000249989 UP000075902 UP000008820 UP000075840 UP000007062
Pfam
PF11901 DUF3421
ProteinModelPortal
H9J0I4
A0A2A4JSG7
A0A2H1V3V6
A0A194PMX8
A0A0N1PHN4
A0A0L7LU66
+ More
A0A212ES78 A0A1E1W9M6 A0A067QXL4 A0A0M8ZRL2 A0A154PJB8 A0A2J7QYZ9 A0A0L7R8N2 F4WZX8 A0A2P8YV38 A0A158NGP6 V9IJU4 A0A151IFH0 T1HED6 A0A2A3E3U6 A0A195FFA6 Q4PKR8 A0A088A6G6 A0A1B6E9E8 A0A151XAE5 E2BD97 A0A1S3D5F2 E9ILB7 D6WN71 A0A1Y1M100 E1ZV65 A0A0T6BG16 N6UFY3 A0A2R7VV21 A0A1W4XIY9 A0A1W4XJX4 B4IHC5 B4QD58 B3NR96 A0A1W4V169 B4PBZ2 A0A0B4LGL6 P10379 A0A1L8DXA3 A0A0K8WAB7 B3MCD4 A0A0A1XP79 A0A1B0CIB2 A0A034VPI8 A0A1A9WV60 A0A3B0JJ97 B4MPM2 A0A3B0J1G1 B4GI31 Q28Z91 B4LP61 W8AJI0 A0A1A9Y731 A0A1I8MTT6 T1PFY7 A0A1B0BGM9 A0A1I8NMY9 A0A1B0FPQ9 A0A1A9Z5C1 B4LA33 A0A1A9VVQ2 A0A084VIF9 A0A0M4EF47 A0A336MUV1 A0A182QP96 A0A182N2H9 A0A336M9W1 A0A0N8DIT6 A0A0P6JJ41 A0A164X310 A0A0P5LK76 A0A182R715 A0A182IJB7 A0A0P6GED9 A0A023ET71 A0A182GZ10 A0A182TRJ3 Q17N01 A0A1Y9GKG9 Q7PZ21
A0A212ES78 A0A1E1W9M6 A0A067QXL4 A0A0M8ZRL2 A0A154PJB8 A0A2J7QYZ9 A0A0L7R8N2 F4WZX8 A0A2P8YV38 A0A158NGP6 V9IJU4 A0A151IFH0 T1HED6 A0A2A3E3U6 A0A195FFA6 Q4PKR8 A0A088A6G6 A0A1B6E9E8 A0A151XAE5 E2BD97 A0A1S3D5F2 E9ILB7 D6WN71 A0A1Y1M100 E1ZV65 A0A0T6BG16 N6UFY3 A0A2R7VV21 A0A1W4XIY9 A0A1W4XJX4 B4IHC5 B4QD58 B3NR96 A0A1W4V169 B4PBZ2 A0A0B4LGL6 P10379 A0A1L8DXA3 A0A0K8WAB7 B3MCD4 A0A0A1XP79 A0A1B0CIB2 A0A034VPI8 A0A1A9WV60 A0A3B0JJ97 B4MPM2 A0A3B0J1G1 B4GI31 Q28Z91 B4LP61 W8AJI0 A0A1A9Y731 A0A1I8MTT6 T1PFY7 A0A1B0BGM9 A0A1I8NMY9 A0A1B0FPQ9 A0A1A9Z5C1 B4LA33 A0A1A9VVQ2 A0A084VIF9 A0A0M4EF47 A0A336MUV1 A0A182QP96 A0A182N2H9 A0A336M9W1 A0A0N8DIT6 A0A0P6JJ41 A0A164X310 A0A0P5LK76 A0A182R715 A0A182IJB7 A0A0P6GED9 A0A023ET71 A0A182GZ10 A0A182TRJ3 Q17N01 A0A1Y9GKG9 Q7PZ21
Ontologies
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 27,
Likelihood: 0.989102
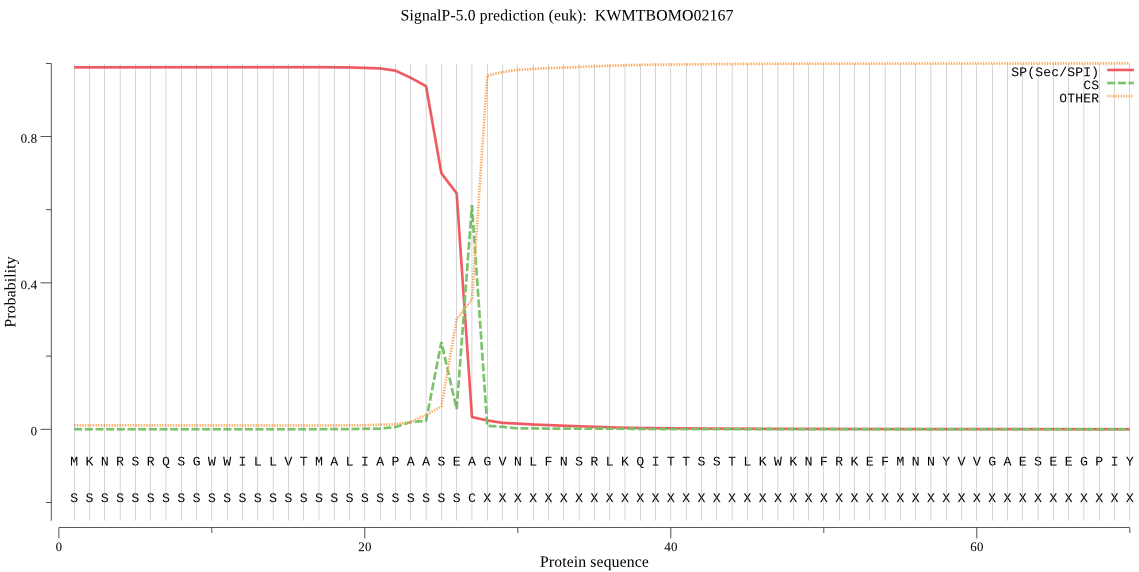
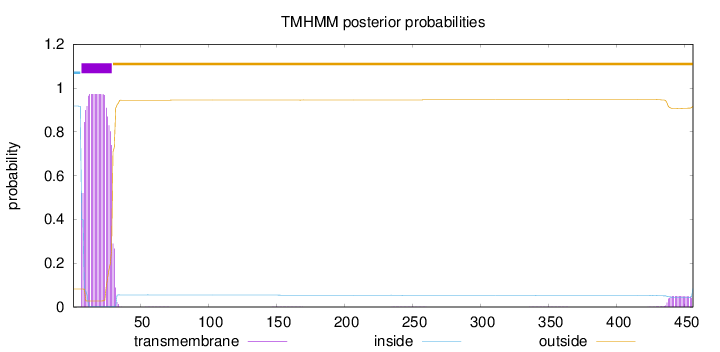
Length:
456
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.11206
Exp number, first 60 AAs:
21.1596
Total prob of N-in:
0.91827
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 456
Population Genetic Test Statistics
Pi
193.02198
Theta
200.706311
Tajima's D
-0.892844
CLR
1019.627226
CSRT
0.15969201539923
Interpretation
Uncertain
