Pre Gene Modal
BGIBMGA003020
Annotation
PREDICTED:_uncharacterized_protein_LOC101745835_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.755 Nuclear Reliability : 1.089
Sequence
CDS
ATGACCCGTACGGTTAAAAAACTGGAAGTTCCCAGACCTCTGAAATCGACGACCAGTTCTGCTCATAACCGTAATTCTACATTGGACGTCAAAATAAACTCCGTTGATGACCTGATAGTACTAGCAGAAGCTGTGGCATACCAAATGATGCAAGGAAATTATGACCGTGCTCTCCAAAGCAATGTTGCCACAATGTACTCGAATCTTAAACTATATGGAGCTCAACTCGAGGTCCTCTACAAAGATTTTCTTGATAGATACTTTGTTGTGTTCCGCAATGGCTCACAAGATGAGCGCCTTGATAAGAAGACTAGATTGCACCTTCTTGAATTGATCGAGTTGAGAGCTAAGTTGTGGCAAGGTTCCGACTATATGAGTCAATATTATAGGCACCGTGGTACTCATGCTGAGCCTTTGCTGGTACCAACAATGGAGGGAACAGGATCAGGTGGGTCTGTGTCGCCTACGCTGGCTGCGCCGGCCGAGCCCCCTGCTATGCTCGGTCCTGGGGAGGTCATCAAACCCTCAGGAAAATTCCCCAAGCCCACCAAAATCCCGGGCAAGAATTACTCCAAAGATGAGGTTGTCATCAGGAATGCTGACTCTGGAAAAGTGATGGGTATTAAGGGCAGGAGGGTGCACATGATCGAGGAGCTAAGTGACACGATCATATCGTTTCAGCGAGTGAGTCCGGGAGCCAAGGAGCGACTAGTACAGATCACTGGACCGAATGAGGAAAATGTCAATCACGCTAAACATCTGATTGGCGACACGATCCGTCGCAACGCGTCCCCGGTGCGACTGGAGGGGTCACTAGAGGCGCCCCGCTCCCCCTCCCGCGCCTCGCTGGACTCTGCCGCCTCTGACGAACCGCCGCGTAACCGAGAGAAATCTCCTCACAACTCGAGAGCTCTGCTTCACAGTTTCTCTACGAATGATGCTGCACTTGGTGAATATAAGTACACTGTGACTTTCGGACAACATTCCATTAAGATCACCGGTAATAACCTGGATCTTGTGAAGACGGCGAAACTCGTGCTGGACGAGTACTTTGAGAGCGCGGGAGCACTGGAGGCCCTCGGCGCCGGCTCCGGGGACTTCTTCACTCTCTCGCAGCGGCCCGGCGCCGCGCTGCTGCTGCGGGACGAAGACGCCGCGCTGCACGACGACGACGTGTTCGTCCCCGCGCCCGCCGCCCTCCCCGCCGACGCAGCCGCCGCCGACGGAGACCTCACGCCGCGCCGACCGCGCTTCTCACGCGCCACCAGCACTGACAAGAACCAAGGTGTAGGCGGCGGCGGGCGGCAGACGTACACGTACGAGACGCTGGTCCAGTACGCGGACTCGCCGCTCAGCAAGCTGCCGCCCGCGGGGCTGGCCGGCTTCCCGCCCGAGCTCGCGCGCAAGAGCTGCCTGGAGTTCGACAGCGCGTCCTACTTGAAGAAGGTCCGCGCGGCGGGGGGCACGTCCGTGGCCACAGTGGACGCGCCCGACTCGCCGGAGCCGGAATAA
Protein
MTRTVKKLEVPRPLKSTTSSAHNRNSTLDVKINSVDDLIVLAEAVAYQMMQGNYDRALQSNVATMYSNLKLYGAQLEVLYKDFLDRYFVVFRNGSQDERLDKKTRLHLLELIELRAKLWQGSDYMSQYYRHRGTHAEPLLVPTMEGTGSGGSVSPTLAAPAEPPAMLGPGEVIKPSGKFPKPTKIPGKNYSKDEVVIRNADSGKVMGIKGRRVHMIEELSDTIISFQRVSPGAKERLVQITGPNEENVNHAKHLIGDTIRRNASPVRLEGSLEAPRSPSRASLDSAASDEPPRNREKSPHNSRALLHSFSTNDAALGEYKYTVTFGQHSIKITGNNLDLVKTAKLVLDEYFESAGALEALGAGSGDFFTLSQRPGAALLLRDEDAALHDDDVFVPAPAALPADAAAADGDLTPRRPRFSRATSTDKNQGVGGGGRQTYTYETLVQYADSPLSKLPPAGLAGFPPELARKSCLEFDSASYLKKVRAAGGTSVATVDAPDSPEPE
Summary
Uniprot
H9J0I3
A0A2A4JS86
A0A194PU62
A0A0N1IFF9
A0A2H1X2Q2
A0A212ES64
+ More
A0A2A4JRU0 A0A2A4JQK1 A0A0L7LUU6 A0A1E1WEK0 A0A1E1W1W0 A0A1E1WSA1 A0A1E1WK79 A0A2J7R0J9 A0A2J7R0J4 A0A2J7R0J5 E2BJN8 E2AAT5 A0A026X386 E9ICW2 A0A2P8ZE76 V9I9B7 V9IAN7 A0A088A1E6 A0A154P4X0 A0A0L7QVR0 A0A195FV26 A0A195CWU5 F4W6S0 A0A195BRT7 A0A151XJZ7 A0A158NS89 A0A151J7Z8 A0A0C9QMZ0 A0A2J7R0I8 K7IQU6 A0A1L8DR97 A0A1L8DR40 A0A232FBK1 A0A1L8DRC0 A0A2J7R0J8 A0A2J7R0J6 A0A1B6CC99 A0A1B6D1H2 D3TP11 A0A0P6J0A5 A0A067QKA2 U5EN51 V9I9E0 E0VVE3 A0A1A9VXT0 A0A2A3EIQ3 V9IBM2 A0A336L592 A0A336MN74 A0A1B0FLU6 T1PG10 A0A1I8PQB4 A0A1I8PQ68 A0A1I8MWA7 T1PKB1 A0A1I8PQC4 A0A034VA17 A0A0K8VSS1 A0A1I8MWB1 W8BKY3 A0A1L8DR14 A0A1J1I6Y7
A0A2A4JRU0 A0A2A4JQK1 A0A0L7LUU6 A0A1E1WEK0 A0A1E1W1W0 A0A1E1WSA1 A0A1E1WK79 A0A2J7R0J9 A0A2J7R0J4 A0A2J7R0J5 E2BJN8 E2AAT5 A0A026X386 E9ICW2 A0A2P8ZE76 V9I9B7 V9IAN7 A0A088A1E6 A0A154P4X0 A0A0L7QVR0 A0A195FV26 A0A195CWU5 F4W6S0 A0A195BRT7 A0A151XJZ7 A0A158NS89 A0A151J7Z8 A0A0C9QMZ0 A0A2J7R0I8 K7IQU6 A0A1L8DR97 A0A1L8DR40 A0A232FBK1 A0A1L8DRC0 A0A2J7R0J8 A0A2J7R0J6 A0A1B6CC99 A0A1B6D1H2 D3TP11 A0A0P6J0A5 A0A067QKA2 U5EN51 V9I9E0 E0VVE3 A0A1A9VXT0 A0A2A3EIQ3 V9IBM2 A0A336L592 A0A336MN74 A0A1B0FLU6 T1PG10 A0A1I8PQB4 A0A1I8PQ68 A0A1I8MWA7 T1PKB1 A0A1I8PQC4 A0A034VA17 A0A0K8VSS1 A0A1I8MWB1 W8BKY3 A0A1L8DR14 A0A1J1I6Y7
Pubmed
EMBL
BABH01010395
BABH01010396
NWSH01000765
PCG74333.1
KQ459598
KPI94675.1
+ More
KQ460709 KPJ12732.1 ODYU01012543 SOQ58924.1 AGBW02012845 OWR44346.1 PCG74334.1 PCG74335.1 JTDY01000079 KOB78991.1 GDQN01005675 JAT85379.1 GDQN01010079 JAT80975.1 GDQN01001131 JAT89923.1 GDQN01003669 JAT87385.1 NEVH01008240 PNF34362.1 PNF34360.1 PNF34361.1 GL448604 EFN84096.1 GL438172 EFN69464.1 KK107019 EZA62750.1 GL762419 EFZ21585.1 PYGN01000083 PSN54796.1 JR037309 AEY57703.1 JR037308 AEY57702.1 KQ434819 KZC06892.1 KQ414722 KOC62708.1 KQ981241 KYN44313.1 KQ977171 KYN05143.1 GL887726 EGI70082.1 KQ976421 KYM89077.1 KQ982045 KYQ60742.1 ADTU01024718 ADTU01024719 ADTU01024720 ADTU01024721 KQ979602 KYN20522.1 GBYB01004914 JAG74681.1 PNF34348.1 GFDF01005153 JAV08931.1 GFDF01005151 JAV08933.1 NNAY01000461 OXU28211.1 GFDF01005157 JAV08927.1 PNF34354.1 PNF34358.1 GEDC01026353 JAS10945.1 GEDC01017774 JAS19524.1 EZ423163 ADD19439.1 GDUN01000446 JAN95473.1 KK853343 KDR08285.1 GANO01004261 JAB55610.1 JR037311 AEY57705.1 DS235811 EEB17349.1 KZ288250 PBC31079.1 JR037310 AEY57704.1 UFQS01001772 UFQT01001772 SSX12301.1 SSX31752.1 UFQT01001810 SSX31862.1 CCAG010004762 KA647609 AFP62238.1 KA649134 AFP63763.1 GAKP01020554 JAC38398.1 GDHF01010426 JAI41888.1 GAMC01008912 JAB97643.1 GFDF01005156 JAV08928.1 CVRI01000038 CRK94185.1
KQ460709 KPJ12732.1 ODYU01012543 SOQ58924.1 AGBW02012845 OWR44346.1 PCG74334.1 PCG74335.1 JTDY01000079 KOB78991.1 GDQN01005675 JAT85379.1 GDQN01010079 JAT80975.1 GDQN01001131 JAT89923.1 GDQN01003669 JAT87385.1 NEVH01008240 PNF34362.1 PNF34360.1 PNF34361.1 GL448604 EFN84096.1 GL438172 EFN69464.1 KK107019 EZA62750.1 GL762419 EFZ21585.1 PYGN01000083 PSN54796.1 JR037309 AEY57703.1 JR037308 AEY57702.1 KQ434819 KZC06892.1 KQ414722 KOC62708.1 KQ981241 KYN44313.1 KQ977171 KYN05143.1 GL887726 EGI70082.1 KQ976421 KYM89077.1 KQ982045 KYQ60742.1 ADTU01024718 ADTU01024719 ADTU01024720 ADTU01024721 KQ979602 KYN20522.1 GBYB01004914 JAG74681.1 PNF34348.1 GFDF01005153 JAV08931.1 GFDF01005151 JAV08933.1 NNAY01000461 OXU28211.1 GFDF01005157 JAV08927.1 PNF34354.1 PNF34358.1 GEDC01026353 JAS10945.1 GEDC01017774 JAS19524.1 EZ423163 ADD19439.1 GDUN01000446 JAN95473.1 KK853343 KDR08285.1 GANO01004261 JAB55610.1 JR037311 AEY57705.1 DS235811 EEB17349.1 KZ288250 PBC31079.1 JR037310 AEY57704.1 UFQS01001772 UFQT01001772 SSX12301.1 SSX31752.1 UFQT01001810 SSX31862.1 CCAG010004762 KA647609 AFP62238.1 KA649134 AFP63763.1 GAKP01020554 JAC38398.1 GDHF01010426 JAI41888.1 GAMC01008912 JAB97643.1 GFDF01005156 JAV08928.1 CVRI01000038 CRK94185.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000235965 UP000008237 UP000000311 UP000053097 UP000245037 UP000005203 UP000076502 UP000053825 UP000078541 UP000078542 UP000007755 UP000078540 UP000075809 UP000005205 UP000078492 UP000002358 UP000215335 UP000027135 UP000009046 UP000078200 UP000242457 UP000092444 UP000095301 UP000095300 UP000183832
UP000235965 UP000008237 UP000000311 UP000053097 UP000245037 UP000005203 UP000076502 UP000053825 UP000078541 UP000078542 UP000007755 UP000078540 UP000075809 UP000005205 UP000078492 UP000002358 UP000215335 UP000027135 UP000009046 UP000078200 UP000242457 UP000092444 UP000095301 UP000095300 UP000183832
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J0I3
A0A2A4JS86
A0A194PU62
A0A0N1IFF9
A0A2H1X2Q2
A0A212ES64
+ More
A0A2A4JRU0 A0A2A4JQK1 A0A0L7LUU6 A0A1E1WEK0 A0A1E1W1W0 A0A1E1WSA1 A0A1E1WK79 A0A2J7R0J9 A0A2J7R0J4 A0A2J7R0J5 E2BJN8 E2AAT5 A0A026X386 E9ICW2 A0A2P8ZE76 V9I9B7 V9IAN7 A0A088A1E6 A0A154P4X0 A0A0L7QVR0 A0A195FV26 A0A195CWU5 F4W6S0 A0A195BRT7 A0A151XJZ7 A0A158NS89 A0A151J7Z8 A0A0C9QMZ0 A0A2J7R0I8 K7IQU6 A0A1L8DR97 A0A1L8DR40 A0A232FBK1 A0A1L8DRC0 A0A2J7R0J8 A0A2J7R0J6 A0A1B6CC99 A0A1B6D1H2 D3TP11 A0A0P6J0A5 A0A067QKA2 U5EN51 V9I9E0 E0VVE3 A0A1A9VXT0 A0A2A3EIQ3 V9IBM2 A0A336L592 A0A336MN74 A0A1B0FLU6 T1PG10 A0A1I8PQB4 A0A1I8PQ68 A0A1I8MWA7 T1PKB1 A0A1I8PQC4 A0A034VA17 A0A0K8VSS1 A0A1I8MWB1 W8BKY3 A0A1L8DR14 A0A1J1I6Y7
A0A2A4JRU0 A0A2A4JQK1 A0A0L7LUU6 A0A1E1WEK0 A0A1E1W1W0 A0A1E1WSA1 A0A1E1WK79 A0A2J7R0J9 A0A2J7R0J4 A0A2J7R0J5 E2BJN8 E2AAT5 A0A026X386 E9ICW2 A0A2P8ZE76 V9I9B7 V9IAN7 A0A088A1E6 A0A154P4X0 A0A0L7QVR0 A0A195FV26 A0A195CWU5 F4W6S0 A0A195BRT7 A0A151XJZ7 A0A158NS89 A0A151J7Z8 A0A0C9QMZ0 A0A2J7R0I8 K7IQU6 A0A1L8DR97 A0A1L8DR40 A0A232FBK1 A0A1L8DRC0 A0A2J7R0J8 A0A2J7R0J6 A0A1B6CC99 A0A1B6D1H2 D3TP11 A0A0P6J0A5 A0A067QKA2 U5EN51 V9I9E0 E0VVE3 A0A1A9VXT0 A0A2A3EIQ3 V9IBM2 A0A336L592 A0A336MN74 A0A1B0FLU6 T1PG10 A0A1I8PQB4 A0A1I8PQ68 A0A1I8MWA7 T1PKB1 A0A1I8PQC4 A0A034VA17 A0A0K8VSS1 A0A1I8MWB1 W8BKY3 A0A1L8DR14 A0A1J1I6Y7
PDB
4JHK
E-value=0.00623246,
Score=94
Ontologies
GO
PANTHER
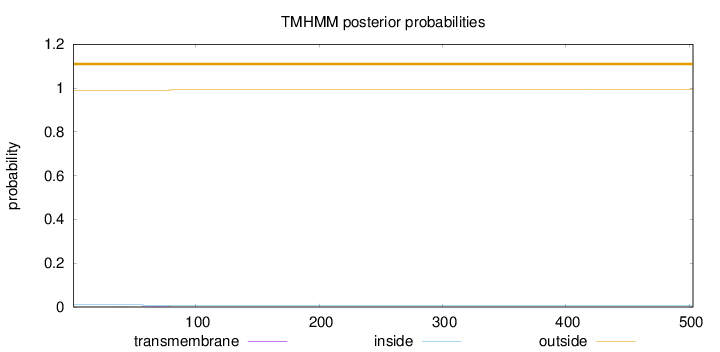
Topology
Length:
503
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0580800000000002
Exp number, first 60 AAs:
0.01324
Total prob of N-in:
0.01049
outside
1 - 503
Population Genetic Test Statistics
Pi
200.25047
Theta
196.554702
Tajima's D
0.172462
CLR
0.230368
CSRT
0.419379031048448
Interpretation
Uncertain
