Gene
KWMTBOMO02161
Pre Gene Modal
BGIBMGA003017
Annotation
PREDICTED:_uncharacterized_protein_LOC101740606_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.942
Sequence
CDS
ATGCAATCGTTAAACGACTTGAACAACGACATGAATTCAAACATTATAATTGTATTATTCTGGGTAAATCAATTTATGTCTTTGAGTAACACAGAAGTACTAACTATTTCGACTAAAGGTTGTACGATACCTTCTTTTGCTAATAACGAGAAAACCATTATAGCTTTGAATGAATCTAAGATTAATTTTAGTTGCCCGAGTTTTCGTCGATTGATCGATGCCAACAAAACGCACATCTGGGTTATTAAAGACGCCTTTGAAGAATTTAATATTTGGAATGAAAATAGTTTTTACTGTTGCTACAGAAGCTTTTCTGCTATAAACAACATATCTAATATCGTATACAAAGACTGTAGGCAATTTTCAAAAATCATTAAGGCGGAAGATGACTTTGTGCGAATACAATGTTATTATGAAGACGACAGAGTATTCGAAGATTTTTTTTTATTCCACATTGAAGATGCAACCAAACAACATGCGCTGATTGACAAAAACCTGTTCAACGTGATAATTATTGGTTTAGAAAGTGTGTCGAGAATGAATTTTCATCGCACAATGAGATTAACATCGAAATATTTAAAAGACCACGGAGCCGTAGAATTCTTTGGTTACAACAAAGTGGGTGATAATTCTTTTCCTAATTTATTTCCCGTTTTGACAGGAAAATCTTTTAAGTACCTTAAAAGAAAACTAAGAAATGAATTTGAACAATGGAATTTTATTTGGGATGATTATAAAAAAGCAGGATTTAATACTGCAATGGGATCAGATAGTATCGCTGGTTTACTCGGATCTTACGAGTATAATTGGGAAAAAATTCCAACAGACAACTATTTACAACCATTTATGTTCGAAGCGAGGCATATGTTCCAGGACAAACAGTACAACTACCACAAGTGCCTAGGTGATAAATTACTTTACAAGATACTTCTGGATTACATCAGTGATATAACTATTAAGCTAAAATCAGTAAGATTATTTGGTTTGTTTTGGGAAGAATCTATTAGTCATGACGATCTCAAATTACCTCATATTATGGATGAAGATTACCTAAATCTTATGAAAAAAATTGAATTCATCGGTAGCTTACGTAATACTGTGATCATTTTCTTCAGTGATCATGGAAAGAGATGGGGAAAGTTTGTTGAAACAAAACAAGGCCGTTTAGAAGAACGTCTACCACTTCTTACTATTATGCTACCTGAAACTTTTAAACGACGTTACACGTTAGCCTTCAAAAACTTGCAACAGAATAGTCGTGGCTTAACAACTCCGTTTGATTTCCACGAAACCCTGTTGGACCTTTTGAACATAACTAAATTAGAAAACCATGTTATTCGTGAAAGAATGAGTGTTTCAAATCAAAATGATAAAGTATCTAGTTTATTTTTACCTATAAGTTCTAAAAGAACATGTAAGGATGTAGGCATACTGCAACACTGGTGTTCTTGCTACAACGGCAGGAGTTTACGGACAGATTCAAAAGACATGACGAGACCATGTACTTCAATGAACGATCAAGAGAAACCCAAGCGGGAAGAAGAGAAAAAAGTTGGTTTGATCCAGAGATTTAAAGAAATGTACAGGGATTACTGGTATGTGTTACTTCCAGTACACATGGCGACTTCCGCTGTATGGTTCGGTAGCTGCTATTATACTGTTAGGAGTGGGGTGGACGTCATTAGTATTTTAGAGTCCCTAGGAGTCACAGAGACACTCTTAAAACCGCTGAAAGAATCTGGTGCTGGATATTTTGCACTTGCCTTTGCCTTGTATAAATTGGTGACACCTTTGCGGTACACAGTTACAGTTGGTGGAACCACTTATGCTATTAAGAGATTAACGGCACTAGGTTGGATTAAACCAGTTCCATCTAAAGAGAGGCTGAAAGAAATGTTACAAGAAAAAAAAGACTTGCTTCAAGAAAAGAAAGATAATCTTCAAGACCGTCTGCATGAGAGTAAACAACATTATCAAGCACAGATACAGGAAAAACGTACACATATGATGGATGAAATGAGAAGATATAAAGCAGAAATGCGCGAAATGAAAAACAAAGTTAAAAAGATAAAAATAAAATCCAGAGGAAACTGA
Protein
MQSLNDLNNDMNSNIIIVLFWVNQFMSLSNTEVLTISTKGCTIPSFANNEKTIIALNESKINFSCPSFRRLIDANKTHIWVIKDAFEEFNIWNENSFYCCYRSFSAINNISNIVYKDCRQFSKIIKAEDDFVRIQCYYEDDRVFEDFFLFHIEDATKQHALIDKNLFNVIIIGLESVSRMNFHRTMRLTSKYLKDHGAVEFFGYNKVGDNSFPNLFPVLTGKSFKYLKRKLRNEFEQWNFIWDDYKKAGFNTAMGSDSIAGLLGSYEYNWEKIPTDNYLQPFMFEARHMFQDKQYNYHKCLGDKLLYKILLDYISDITIKLKSVRLFGLFWEESISHDDLKLPHIMDEDYLNLMKKIEFIGSLRNTVIIFFSDHGKRWGKFVETKQGRLEERLPLLTIMLPETFKRRYTLAFKNLQQNSRGLTTPFDFHETLLDLLNITKLENHVIRERMSVSNQNDKVSSLFLPISSKRTCKDVGILQHWCSCYNGRSLRTDSKDMTRPCTSMNDQEKPKREEEKKVGLIQRFKEMYRDYWYVLLPVHMATSAVWFGSCYYTVRSGVDVISILESLGVTETLLKPLKESGAGYFALAFALYKLVTPLRYTVTVGGTTYAIKRLTALGWIKPVPSKERLKEMLQEKKDLLQEKKDNLQDRLHESKQHYQAQIQEKRTHMMDEMRRYKAEMREMKNKVKKIKIKSRGN
Summary
Uniprot
H9J0I0
A0A194PU67
A0A0N1PGG2
A0A0N1INZ4
A0A194PMM2
A0A0L7L4W3
+ More
A0A2J7RGB7 A0A2J7RGB5 A0A1B6EXQ9 A0A1B6FI27 A0A195EZZ4 F4WAS6 A0A195BYY4 A0A158P3J4 A0A151XDP9 A0A0L7R113 B0WE52 A0A023EZB6 A0A088AJM9 A0A1I8NM69 A0A2A3E6W0 E9IEB1 A0A151JMM7 A0A0C9RZ88 Q16UC9 J9JY89 A0A0L0BSI2 B3N4L7 A0A1W4W5S4 E1ZX72 B4JAD7 A0A0J7NCX7 Q9VKK3 M9ND65 A0A182G8U5 B4QAA5 A0A154NY24 Q16KF5 A0A1S4FXP3 B4P166 A0A0J9R0L3 A0A182UV73 Q7Q4B8 A0A182SF93 A0A182U9Z6 A0A182L0R9 B4HX11 A0A1S4GXY1 A0A182XLA1 A0A182ICC3 A0A1A9ZN61 A0A182Y3C5 A0A1A9UXA3 A0A1A9WBJ6 A0A1B0G663 A0A1B0ATM7 A0A1A9Y4Q6 A0A182P8K9 A0A182JUE2 A0A1I8NH34 A0A182QQR7 B4G9T1 A0A182NTU2 A0A0R3NSW2
A0A2J7RGB7 A0A2J7RGB5 A0A1B6EXQ9 A0A1B6FI27 A0A195EZZ4 F4WAS6 A0A195BYY4 A0A158P3J4 A0A151XDP9 A0A0L7R113 B0WE52 A0A023EZB6 A0A088AJM9 A0A1I8NM69 A0A2A3E6W0 E9IEB1 A0A151JMM7 A0A0C9RZ88 Q16UC9 J9JY89 A0A0L0BSI2 B3N4L7 A0A1W4W5S4 E1ZX72 B4JAD7 A0A0J7NCX7 Q9VKK3 M9ND65 A0A182G8U5 B4QAA5 A0A154NY24 Q16KF5 A0A1S4FXP3 B4P166 A0A0J9R0L3 A0A182UV73 Q7Q4B8 A0A182SF93 A0A182U9Z6 A0A182L0R9 B4HX11 A0A1S4GXY1 A0A182XLA1 A0A182ICC3 A0A1A9ZN61 A0A182Y3C5 A0A1A9UXA3 A0A1A9WBJ6 A0A1B0G663 A0A1B0ATM7 A0A1A9Y4Q6 A0A182P8K9 A0A182JUE2 A0A1I8NH34 A0A182QQR7 B4G9T1 A0A182NTU2 A0A0R3NSW2
Pubmed
EMBL
BABH01010401
BABH01010402
KQ459598
KPI94680.1
KQ460709
KPJ12727.1
+ More
KPJ12726.1 KPI94681.1 JTDY01002938 KOB70455.1 NEVH01004401 PNF39881.1 PNF39876.1 GECZ01027168 JAS42601.1 GECZ01020166 JAS49603.1 KQ981880 KYN33890.1 GL888053 EGI68682.1 KQ976394 KYM93128.1 ADTU01008160 ADTU01008161 KQ982268 KYQ58483.1 KQ414668 KOC64532.1 DS231904 EDS45228.1 GBBI01004135 JAC14577.1 KZ288349 PBC27448.1 GL762576 EFZ21063.1 KQ978910 KYN27535.1 GBYB01013416 JAG83183.1 CH477624 EAT38139.1 ABLF02039889 JRES01001427 KNC22971.1 CH954177 EDV58929.1 GL435030 EFN74162.1 CH916368 EDW03808.1 LBMM01006653 KMQ90425.1 AE014134 AY118291 AAF53064.1 AAM48320.1 AFH03648.1 AHN54345.1 JXUM01152784 KQ571544 KXJ68159.1 CM000361 EDX04660.1 KQ434782 KZC04517.1 CH477961 EAT34767.1 CM000157 EDW88041.1 CM002910 KMY89691.1 AAAB01008964 EAA12425.3 CH480818 EDW52556.1 APCN01000793 CCAG010007078 JXJN01003397 AXCN02000805 CH479180 EDW29111.1 CH379060 KRT04264.1 KRT04265.1
KPJ12726.1 KPI94681.1 JTDY01002938 KOB70455.1 NEVH01004401 PNF39881.1 PNF39876.1 GECZ01027168 JAS42601.1 GECZ01020166 JAS49603.1 KQ981880 KYN33890.1 GL888053 EGI68682.1 KQ976394 KYM93128.1 ADTU01008160 ADTU01008161 KQ982268 KYQ58483.1 KQ414668 KOC64532.1 DS231904 EDS45228.1 GBBI01004135 JAC14577.1 KZ288349 PBC27448.1 GL762576 EFZ21063.1 KQ978910 KYN27535.1 GBYB01013416 JAG83183.1 CH477624 EAT38139.1 ABLF02039889 JRES01001427 KNC22971.1 CH954177 EDV58929.1 GL435030 EFN74162.1 CH916368 EDW03808.1 LBMM01006653 KMQ90425.1 AE014134 AY118291 AAF53064.1 AAM48320.1 AFH03648.1 AHN54345.1 JXUM01152784 KQ571544 KXJ68159.1 CM000361 EDX04660.1 KQ434782 KZC04517.1 CH477961 EAT34767.1 CM000157 EDW88041.1 CM002910 KMY89691.1 AAAB01008964 EAA12425.3 CH480818 EDW52556.1 APCN01000793 CCAG010007078 JXJN01003397 AXCN02000805 CH479180 EDW29111.1 CH379060 KRT04264.1 KRT04265.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000235965
UP000078541
+ More
UP000007755 UP000078540 UP000005205 UP000075809 UP000053825 UP000002320 UP000005203 UP000095300 UP000242457 UP000078492 UP000008820 UP000007819 UP000037069 UP000008711 UP000192221 UP000000311 UP000001070 UP000036403 UP000000803 UP000069940 UP000249989 UP000000304 UP000076502 UP000002282 UP000075903 UP000007062 UP000075901 UP000075902 UP000075882 UP000001292 UP000076407 UP000075840 UP000092445 UP000076408 UP000078200 UP000091820 UP000092444 UP000092460 UP000092443 UP000075885 UP000075881 UP000095301 UP000075886 UP000008744 UP000075884 UP000001819
UP000007755 UP000078540 UP000005205 UP000075809 UP000053825 UP000002320 UP000005203 UP000095300 UP000242457 UP000078492 UP000008820 UP000007819 UP000037069 UP000008711 UP000192221 UP000000311 UP000001070 UP000036403 UP000000803 UP000069940 UP000249989 UP000000304 UP000076502 UP000002282 UP000075903 UP000007062 UP000075901 UP000075902 UP000075882 UP000001292 UP000076407 UP000075840 UP000092445 UP000076408 UP000078200 UP000091820 UP000092444 UP000092460 UP000092443 UP000075885 UP000075881 UP000095301 UP000075886 UP000008744 UP000075884 UP000001819
Interpro
Gene 3D
ProteinModelPortal
H9J0I0
A0A194PU67
A0A0N1PGG2
A0A0N1INZ4
A0A194PMM2
A0A0L7L4W3
+ More
A0A2J7RGB7 A0A2J7RGB5 A0A1B6EXQ9 A0A1B6FI27 A0A195EZZ4 F4WAS6 A0A195BYY4 A0A158P3J4 A0A151XDP9 A0A0L7R113 B0WE52 A0A023EZB6 A0A088AJM9 A0A1I8NM69 A0A2A3E6W0 E9IEB1 A0A151JMM7 A0A0C9RZ88 Q16UC9 J9JY89 A0A0L0BSI2 B3N4L7 A0A1W4W5S4 E1ZX72 B4JAD7 A0A0J7NCX7 Q9VKK3 M9ND65 A0A182G8U5 B4QAA5 A0A154NY24 Q16KF5 A0A1S4FXP3 B4P166 A0A0J9R0L3 A0A182UV73 Q7Q4B8 A0A182SF93 A0A182U9Z6 A0A182L0R9 B4HX11 A0A1S4GXY1 A0A182XLA1 A0A182ICC3 A0A1A9ZN61 A0A182Y3C5 A0A1A9UXA3 A0A1A9WBJ6 A0A1B0G663 A0A1B0ATM7 A0A1A9Y4Q6 A0A182P8K9 A0A182JUE2 A0A1I8NH34 A0A182QQR7 B4G9T1 A0A182NTU2 A0A0R3NSW2
A0A2J7RGB7 A0A2J7RGB5 A0A1B6EXQ9 A0A1B6FI27 A0A195EZZ4 F4WAS6 A0A195BYY4 A0A158P3J4 A0A151XDP9 A0A0L7R113 B0WE52 A0A023EZB6 A0A088AJM9 A0A1I8NM69 A0A2A3E6W0 E9IEB1 A0A151JMM7 A0A0C9RZ88 Q16UC9 J9JY89 A0A0L0BSI2 B3N4L7 A0A1W4W5S4 E1ZX72 B4JAD7 A0A0J7NCX7 Q9VKK3 M9ND65 A0A182G8U5 B4QAA5 A0A154NY24 Q16KF5 A0A1S4FXP3 B4P166 A0A0J9R0L3 A0A182UV73 Q7Q4B8 A0A182SF93 A0A182U9Z6 A0A182L0R9 B4HX11 A0A1S4GXY1 A0A182XLA1 A0A182ICC3 A0A1A9ZN61 A0A182Y3C5 A0A1A9UXA3 A0A1A9WBJ6 A0A1B0G663 A0A1B0ATM7 A0A1A9Y4Q6 A0A182P8K9 A0A182JUE2 A0A1I8NH34 A0A182QQR7 B4G9T1 A0A182NTU2 A0A0R3NSW2
Ontologies
GO
PANTHER
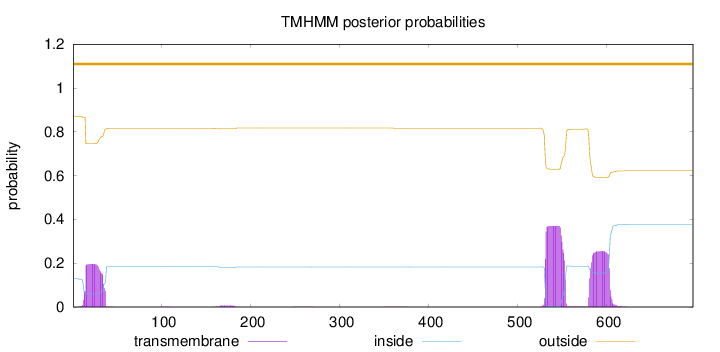
Topology
Length:
697
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
18.07636
Exp number, first 60 AAs:
4.15874
Total prob of N-in:
0.12952
outside
1 - 697
Population Genetic Test Statistics
Pi
168.01123
Theta
179.393407
Tajima's D
-0.398729
CLR
0.535671
CSRT
0.271336433178341
Interpretation
Possibly Positive selection
