Gene
KWMTBOMO02160
Pre Gene Modal
BGIBMGA003016
Annotation
PREDICTED:_uncharacterized_protein_LOC101746350_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.173 PlasmaMembrane Reliability : 1.641
Sequence
CDS
ATGACTGCAGCAGCAAAAGGTGGCGGGAGACCGCGAGCTGTGCCGTTGCGCTTCAAATATTTGTTATTGCTCACAACTATCTTCGGTGGCTGCCTGATCCTCGTGACATTCAACTCACGCATAGAGATGTTCATTATGAACACTGACCCGTCCTGGAGTCATCGCTATGAAATAAGAGATTCTGAAGAAAGTTACACAATTAGAACGGAAGGATGCACGATACCAGGACTCAGGGCGTTTGATGAAAGCATTAAAAAATTCAAAGACTCAATTGAAACGATACAAAATCAAAGTGGAGGATACAATGTAATTGTGATGGGTATTGATGCTATTTCCAGGTTAAATTTCCACAGGACTATGCCTAAGACGCTGGCGTATCTCAAAAAGAAGGGAGGATTGGAGTTCCTTGGCTATAATAAAGTTGGTGACAACACATTTCCAAATTTAATACCTATGCTGTTGGGTATCAAAGATACAGAATTAAAAAAAACGTGTTGGCCGAACACAAAATCTACCTTTGATAATTGTCCATTTATTTGGGACTGGTATAAAGATGCTGGGTACTACACTGCATTTGGTGAAGACAGTGCCAGACTAGGAACATTTAACTACGAAAAACTCGGGTTTGCGGCTTCTCCCACGGATTATTACATTCACACGTTCATGTACGAAGCTGAAATTCACGTGGGGAACAAAAAAGATTTTAACGCTTTCATTTGCATGGGTGATAAATACTTTTATAAGGTTCTCCTAGACTATATAGAAAGTTTAACTCGCACATTGAAAAACGATAAGCTATTCGGGTTTTTTTGGGAAGTCACAATGAGCCACGACTATTTAAATTATCCTATGTTGATGGATGACGATTACGAACAATTTTTGAAACATCTCGATGACAGTAATTACTTAGATGACACGATTTTAATTTTACTTAGTGATCATGGAATTAGATGGGGTGAAATAAGATATACAAAACAAGGTAGATTAGAAGAAAGACTACCCTTGTTCCAAATATTAATACCACGTTCATTTCAAGATAACTTTCCCATGGCTTATGAAAATATTAAAAATAACAGAAAACGTCTCACTACGCCATTTGATTTACATGCAACACTTTCCGATTTGACGAATTTAAATTTCATAAAAGACGAACAAATTATTTTACGAAGCAAAGAAAATTACGGCTCCGATAGAAGTATAAGTCTATTTTTACCAATCCCAAATAATCGAACATGTAATACAGCTGCTATTGATGATCATTGGTGCACATGTCATAAGAGTAATAGGATTTCAATCAATAACAAAGACGTGTACGAGCCCGCCTTCGAACTTGTCAGAAATTTAAATTTGTTATTACAAGATCATTCCCAATGTGTCAAATTGAAGCTTTCGGAGATTCTTGAAGCCACGGAAATGGATGCAGGTGTCCCCGAAGTAGGTGAAGTCGGTTGGCAAGAGATTATGCTGGTGGTACGAACTACTCCCGGAGGAGGAGTCTTTGAAGCAACCCTTCGACGTAACAGTAAACAGGAGTGGTCTCTCGCCGGTACTGTCAGTCGACTTAACCTATACGGAGATCAAAGCCGTTGTGTTCATCATTATCAGTTAAAATTATATTGTTTCTGTGTCTAA
Protein
MTAAAKGGGRPRAVPLRFKYLLLLTTIFGGCLILVTFNSRIEMFIMNTDPSWSHRYEIRDSEESYTIRTEGCTIPGLRAFDESIKKFKDSIETIQNQSGGYNVIVMGIDAISRLNFHRTMPKTLAYLKKKGGLEFLGYNKVGDNTFPNLIPMLLGIKDTELKKTCWPNTKSTFDNCPFIWDWYKDAGYYTAFGEDSARLGTFNYEKLGFAASPTDYYIHTFMYEAEIHVGNKKDFNAFICMGDKYFYKVLLDYIESLTRTLKNDKLFGFFWEVTMSHDYLNYPMLMDDDYEQFLKHLDDSNYLDDTILILLSDHGIRWGEIRYTKQGRLEERLPLFQILIPRSFQDNFPMAYENIKNNRKRLTTPFDLHATLSDLTNLNFIKDEQIILRSKENYGSDRSISLFLPIPNNRTCNTAAIDDHWCTCHKSNRISINNKDVYEPAFELVRNLNLLLQDHSQCVKLKLSEILEATEMDAGVPEVGEVGWQEIMLVVRTTPGGGVFEATLRRNSKQEWSLAGTVSRLNLYGDQSRCVHHYQLKLYCFCV
Summary
Uniprot
H9J0H9
A0A194PMM2
A0A0N1INZ4
A0A2H1WJP3
A0A0L7L4W3
A0A194PU67
+ More
A0A0N1PGG2 A0A212F6Z2 A0A2A4JGG1 A0A1Y1K644 A0A1Y1K197 A0A1B6J9G4 A0A1B6F3L2 A0A1B6FMS1 A0A1B6FI27 A0A1B6EXQ9 A0A2J7RGC8 A0A067RCI7 A0A2J7RGB7 A0A2J7RGB5 T1IG22 A0A084VEC3 A0A023EZB6 A0A1B6C7V4 A0A2P8ZDQ3 A0A182JM29 A0A182NTU2 A0A2P8ZDR0 A0A182P8K9 Q7Q4B8 A0A0C9RZ88 A0A182U9Z6 A0A182XLA1 A0A1S4GXY1 A0A182L0R9 A0A182ICC3 A0A182QQR7 A0A182Y3C5 A0A182UV73 A0A182W6L1 E1ZX72 A0A1W4W5L0 A0A182M1M8 F4WAS6 B4QAA5 A0A158P3J4 A0A026WTI5 B0WE52 A0A3L8DJ23 A0A0L7R113 A0A195BYY4 A0A195EZZ4 A0A2R7WQ16 B4HX11 A0A0J9R0L3 A0A182RMA9 W5J5Y5 A0A151IEK0 Q9VKK3 A0A182G8U5 A0A0N0U383 A0A182F1E8 Q16UC9 M9ND65 B3N4L7 A0A2S2PH97 E2BBA1 A0A1I8NH34 A0A3B0K1J2 A0A151JMM7 B4JAD7 D6WL82 A0A0R3NSW2 A0A151XDP9 A0A1B0CCL8 B4P166 Q29M46 A0A0L0BSI2 A0A1W4W5S4 E9IEB1 A0A0J7NCX7 A0A182JUE2 A0A1I8NM69 B4G9T1 A0A0M4EFJ5 A0A1S4FXP3 Q16KF5 A0A088AJM9 B4LTF3 A0A310SUG3 B4NLQ0 J9JY89 E1ZX71 A0A1A9Y4Q6 A0A1B0ATM7 A0A1A9ZN61 A0A154NY24 A0A1S3D6S8 A0A1B0G663 A0A1A9UXA3
A0A0N1PGG2 A0A212F6Z2 A0A2A4JGG1 A0A1Y1K644 A0A1Y1K197 A0A1B6J9G4 A0A1B6F3L2 A0A1B6FMS1 A0A1B6FI27 A0A1B6EXQ9 A0A2J7RGC8 A0A067RCI7 A0A2J7RGB7 A0A2J7RGB5 T1IG22 A0A084VEC3 A0A023EZB6 A0A1B6C7V4 A0A2P8ZDQ3 A0A182JM29 A0A182NTU2 A0A2P8ZDR0 A0A182P8K9 Q7Q4B8 A0A0C9RZ88 A0A182U9Z6 A0A182XLA1 A0A1S4GXY1 A0A182L0R9 A0A182ICC3 A0A182QQR7 A0A182Y3C5 A0A182UV73 A0A182W6L1 E1ZX72 A0A1W4W5L0 A0A182M1M8 F4WAS6 B4QAA5 A0A158P3J4 A0A026WTI5 B0WE52 A0A3L8DJ23 A0A0L7R113 A0A195BYY4 A0A195EZZ4 A0A2R7WQ16 B4HX11 A0A0J9R0L3 A0A182RMA9 W5J5Y5 A0A151IEK0 Q9VKK3 A0A182G8U5 A0A0N0U383 A0A182F1E8 Q16UC9 M9ND65 B3N4L7 A0A2S2PH97 E2BBA1 A0A1I8NH34 A0A3B0K1J2 A0A151JMM7 B4JAD7 D6WL82 A0A0R3NSW2 A0A151XDP9 A0A1B0CCL8 B4P166 Q29M46 A0A0L0BSI2 A0A1W4W5S4 E9IEB1 A0A0J7NCX7 A0A182JUE2 A0A1I8NM69 B4G9T1 A0A0M4EFJ5 A0A1S4FXP3 Q16KF5 A0A088AJM9 B4LTF3 A0A310SUG3 B4NLQ0 J9JY89 E1ZX71 A0A1A9Y4Q6 A0A1B0ATM7 A0A1A9ZN61 A0A154NY24 A0A1S3D6S8 A0A1B0G663 A0A1A9UXA3
Pubmed
19121390
26354079
26227816
22118469
28004739
24845553
+ More
24438588 25474469 29403074 12364791 20966253 25244985 20798317 21719571 17994087 21347285 24508170 30249741 22936249 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 17510324 25315136 18362917 19820115 15632085 23185243 17550304 26108605 21282665
24438588 25474469 29403074 12364791 20966253 25244985 20798317 21719571 17994087 21347285 24508170 30249741 22936249 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 17510324 25315136 18362917 19820115 15632085 23185243 17550304 26108605 21282665
EMBL
BABH01010402
KQ459598
KPI94681.1
KQ460709
KPJ12726.1
ODYU01009115
+ More
SOQ53305.1 JTDY01002938 KOB70455.1 KPI94680.1 KPJ12727.1 AGBW02009947 OWR49516.1 NWSH01001661 PCG70510.1 GEZM01097358 JAV54257.1 GEZM01097357 JAV54258.1 GECU01011894 JAS95812.1 GECZ01024920 JAS44849.1 GECZ01018315 JAS51454.1 GECZ01020166 JAS49603.1 GECZ01027168 JAS42601.1 NEVH01004401 PNF39878.1 KK852595 KDR20616.1 PNF39881.1 PNF39876.1 ACPB03022605 ATLV01012268 KE524777 KFB36317.1 GBBI01004135 JAC14577.1 GEDC01027715 JAS09583.1 PYGN01000087 PSN54629.1 PSN54628.1 AAAB01008964 EAA12425.3 GBYB01013416 JAG83183.1 APCN01000793 AXCN02000805 GL435030 EFN74162.1 AXCM01001485 GL888053 EGI68682.1 CM000361 EDX04660.1 ADTU01008160 ADTU01008161 KK107109 EZA59268.1 DS231904 EDS45228.1 QOIP01000008 RLU19929.1 KQ414668 KOC64532.1 KQ976394 KYM93128.1 KQ981880 KYN33890.1 KK855154 PTY21180.1 CH480818 EDW52556.1 CM002910 KMY89691.1 ADMH02002157 ETN58209.1 KQ977866 KYM99247.1 AE014134 AY118291 AAF53064.1 AAM48320.1 JXUM01152784 KQ571544 KXJ68159.1 KQ435964 KOX67951.1 CH477624 EAT38139.1 AFH03648.1 AHN54345.1 CH954177 EDV58929.1 GGMR01016184 MBY28803.1 GL446957 EFN87029.1 EFN87031.1 OUUW01000004 SPP79466.1 KQ978910 KYN27535.1 CH916368 EDW03808.1 KQ971343 EFA03493.1 CH379060 KRT04264.1 KRT04265.1 KQ982268 KYQ58483.1 AJWK01006903 CM000157 EDW88041.1 EAL33849.3 JRES01001427 KNC22971.1 GL762576 EFZ21063.1 LBMM01006653 KMQ90425.1 CH479180 EDW29111.1 CP012523 ALC39230.1 CH477961 EAT34767.1 CH940649 EDW63923.1 KQ760158 OAD61696.1 CH964274 EDW85289.1 ABLF02039889 EFN74161.1 JXJN01003397 KQ434782 KZC04517.1 CCAG010007078
SOQ53305.1 JTDY01002938 KOB70455.1 KPI94680.1 KPJ12727.1 AGBW02009947 OWR49516.1 NWSH01001661 PCG70510.1 GEZM01097358 JAV54257.1 GEZM01097357 JAV54258.1 GECU01011894 JAS95812.1 GECZ01024920 JAS44849.1 GECZ01018315 JAS51454.1 GECZ01020166 JAS49603.1 GECZ01027168 JAS42601.1 NEVH01004401 PNF39878.1 KK852595 KDR20616.1 PNF39881.1 PNF39876.1 ACPB03022605 ATLV01012268 KE524777 KFB36317.1 GBBI01004135 JAC14577.1 GEDC01027715 JAS09583.1 PYGN01000087 PSN54629.1 PSN54628.1 AAAB01008964 EAA12425.3 GBYB01013416 JAG83183.1 APCN01000793 AXCN02000805 GL435030 EFN74162.1 AXCM01001485 GL888053 EGI68682.1 CM000361 EDX04660.1 ADTU01008160 ADTU01008161 KK107109 EZA59268.1 DS231904 EDS45228.1 QOIP01000008 RLU19929.1 KQ414668 KOC64532.1 KQ976394 KYM93128.1 KQ981880 KYN33890.1 KK855154 PTY21180.1 CH480818 EDW52556.1 CM002910 KMY89691.1 ADMH02002157 ETN58209.1 KQ977866 KYM99247.1 AE014134 AY118291 AAF53064.1 AAM48320.1 JXUM01152784 KQ571544 KXJ68159.1 KQ435964 KOX67951.1 CH477624 EAT38139.1 AFH03648.1 AHN54345.1 CH954177 EDV58929.1 GGMR01016184 MBY28803.1 GL446957 EFN87029.1 EFN87031.1 OUUW01000004 SPP79466.1 KQ978910 KYN27535.1 CH916368 EDW03808.1 KQ971343 EFA03493.1 CH379060 KRT04264.1 KRT04265.1 KQ982268 KYQ58483.1 AJWK01006903 CM000157 EDW88041.1 EAL33849.3 JRES01001427 KNC22971.1 GL762576 EFZ21063.1 LBMM01006653 KMQ90425.1 CH479180 EDW29111.1 CP012523 ALC39230.1 CH477961 EAT34767.1 CH940649 EDW63923.1 KQ760158 OAD61696.1 CH964274 EDW85289.1 ABLF02039889 EFN74161.1 JXJN01003397 KQ434782 KZC04517.1 CCAG010007078
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000218220
+ More
UP000235965 UP000027135 UP000015103 UP000030765 UP000245037 UP000075880 UP000075884 UP000075885 UP000007062 UP000075902 UP000076407 UP000075882 UP000075840 UP000075886 UP000076408 UP000075903 UP000075920 UP000000311 UP000192223 UP000075883 UP000007755 UP000000304 UP000005205 UP000053097 UP000002320 UP000279307 UP000053825 UP000078540 UP000078541 UP000001292 UP000075900 UP000000673 UP000078542 UP000000803 UP000069940 UP000249989 UP000053105 UP000069272 UP000008820 UP000008711 UP000008237 UP000095301 UP000268350 UP000078492 UP000001070 UP000007266 UP000001819 UP000075809 UP000092461 UP000002282 UP000037069 UP000192221 UP000036403 UP000075881 UP000095300 UP000008744 UP000092553 UP000005203 UP000008792 UP000007798 UP000007819 UP000092443 UP000092460 UP000092445 UP000076502 UP000079169 UP000092444 UP000078200
UP000235965 UP000027135 UP000015103 UP000030765 UP000245037 UP000075880 UP000075884 UP000075885 UP000007062 UP000075902 UP000076407 UP000075882 UP000075840 UP000075886 UP000076408 UP000075903 UP000075920 UP000000311 UP000192223 UP000075883 UP000007755 UP000000304 UP000005205 UP000053097 UP000002320 UP000279307 UP000053825 UP000078540 UP000078541 UP000001292 UP000075900 UP000000673 UP000078542 UP000000803 UP000069940 UP000249989 UP000053105 UP000069272 UP000008820 UP000008711 UP000008237 UP000095301 UP000268350 UP000078492 UP000001070 UP000007266 UP000001819 UP000075809 UP000092461 UP000002282 UP000037069 UP000192221 UP000036403 UP000075881 UP000095300 UP000008744 UP000092553 UP000005203 UP000008792 UP000007798 UP000007819 UP000092443 UP000092460 UP000092445 UP000076502 UP000079169 UP000092444 UP000078200
Interpro
Gene 3D
ProteinModelPortal
H9J0H9
A0A194PMM2
A0A0N1INZ4
A0A2H1WJP3
A0A0L7L4W3
A0A194PU67
+ More
A0A0N1PGG2 A0A212F6Z2 A0A2A4JGG1 A0A1Y1K644 A0A1Y1K197 A0A1B6J9G4 A0A1B6F3L2 A0A1B6FMS1 A0A1B6FI27 A0A1B6EXQ9 A0A2J7RGC8 A0A067RCI7 A0A2J7RGB7 A0A2J7RGB5 T1IG22 A0A084VEC3 A0A023EZB6 A0A1B6C7V4 A0A2P8ZDQ3 A0A182JM29 A0A182NTU2 A0A2P8ZDR0 A0A182P8K9 Q7Q4B8 A0A0C9RZ88 A0A182U9Z6 A0A182XLA1 A0A1S4GXY1 A0A182L0R9 A0A182ICC3 A0A182QQR7 A0A182Y3C5 A0A182UV73 A0A182W6L1 E1ZX72 A0A1W4W5L0 A0A182M1M8 F4WAS6 B4QAA5 A0A158P3J4 A0A026WTI5 B0WE52 A0A3L8DJ23 A0A0L7R113 A0A195BYY4 A0A195EZZ4 A0A2R7WQ16 B4HX11 A0A0J9R0L3 A0A182RMA9 W5J5Y5 A0A151IEK0 Q9VKK3 A0A182G8U5 A0A0N0U383 A0A182F1E8 Q16UC9 M9ND65 B3N4L7 A0A2S2PH97 E2BBA1 A0A1I8NH34 A0A3B0K1J2 A0A151JMM7 B4JAD7 D6WL82 A0A0R3NSW2 A0A151XDP9 A0A1B0CCL8 B4P166 Q29M46 A0A0L0BSI2 A0A1W4W5S4 E9IEB1 A0A0J7NCX7 A0A182JUE2 A0A1I8NM69 B4G9T1 A0A0M4EFJ5 A0A1S4FXP3 Q16KF5 A0A088AJM9 B4LTF3 A0A310SUG3 B4NLQ0 J9JY89 E1ZX71 A0A1A9Y4Q6 A0A1B0ATM7 A0A1A9ZN61 A0A154NY24 A0A1S3D6S8 A0A1B0G663 A0A1A9UXA3
A0A0N1PGG2 A0A212F6Z2 A0A2A4JGG1 A0A1Y1K644 A0A1Y1K197 A0A1B6J9G4 A0A1B6F3L2 A0A1B6FMS1 A0A1B6FI27 A0A1B6EXQ9 A0A2J7RGC8 A0A067RCI7 A0A2J7RGB7 A0A2J7RGB5 T1IG22 A0A084VEC3 A0A023EZB6 A0A1B6C7V4 A0A2P8ZDQ3 A0A182JM29 A0A182NTU2 A0A2P8ZDR0 A0A182P8K9 Q7Q4B8 A0A0C9RZ88 A0A182U9Z6 A0A182XLA1 A0A1S4GXY1 A0A182L0R9 A0A182ICC3 A0A182QQR7 A0A182Y3C5 A0A182UV73 A0A182W6L1 E1ZX72 A0A1W4W5L0 A0A182M1M8 F4WAS6 B4QAA5 A0A158P3J4 A0A026WTI5 B0WE52 A0A3L8DJ23 A0A0L7R113 A0A195BYY4 A0A195EZZ4 A0A2R7WQ16 B4HX11 A0A0J9R0L3 A0A182RMA9 W5J5Y5 A0A151IEK0 Q9VKK3 A0A182G8U5 A0A0N0U383 A0A182F1E8 Q16UC9 M9ND65 B3N4L7 A0A2S2PH97 E2BBA1 A0A1I8NH34 A0A3B0K1J2 A0A151JMM7 B4JAD7 D6WL82 A0A0R3NSW2 A0A151XDP9 A0A1B0CCL8 B4P166 Q29M46 A0A0L0BSI2 A0A1W4W5S4 E9IEB1 A0A0J7NCX7 A0A182JUE2 A0A1I8NM69 B4G9T1 A0A0M4EFJ5 A0A1S4FXP3 Q16KF5 A0A088AJM9 B4LTF3 A0A310SUG3 B4NLQ0 J9JY89 E1ZX71 A0A1A9Y4Q6 A0A1B0ATM7 A0A1A9ZN61 A0A154NY24 A0A1S3D6S8 A0A1B0G663 A0A1A9UXA3
Ontologies
GO
PANTHER
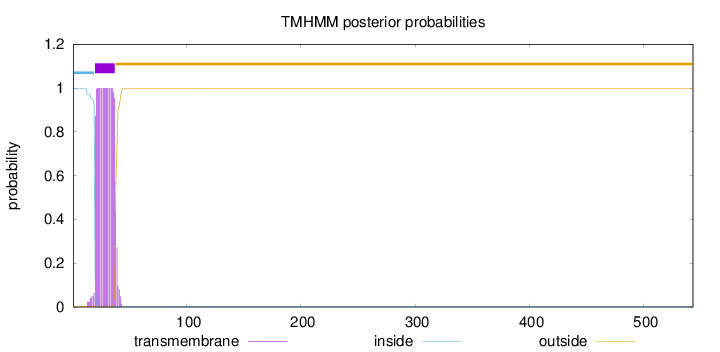
Topology
Length:
543
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.05686
Exp number, first 60 AAs:
19.0318
Total prob of N-in:
0.99711
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 37
outside
38 - 543
Population Genetic Test Statistics
Pi
150.745026
Theta
161.485629
Tajima's D
-0.651472
CLR
1.016747
CSRT
0.208089595520224
Interpretation
Uncertain
