Gene
KWMTBOMO02156
Pre Gene Modal
BGIBMGA003013
Annotation
PREDICTED:_whirlin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.972 Mitochondrial Reliability : 1.41
Sequence
CDS
ATGTCTCGGAGCTTCGACATGAAGGAGGAAGGCAATGCTCTGATTGATGATCGAGGAGGAATTCGAAGGCATCAGTCAATGCAGCGTCTACAGCACGGAGATCAGCAGACCGATGAAAGTCGAGCAGCTAGGGTCGTATCGGACCACCACGGAAACTTACACATCACCGTTAAAAAGACTAAACCAATACTCGGCATAGCGATAGAAGGCGGCGCTAACACCAAACATCCACTACCTCGAATTATAAATATACATGAACATGGAGCCGCATACGAGGCTGGAGGACTAGAGGTGGGCCAGCTGATTTTGGCCGTAGATGGACACCGCGTGGAGGGCATGCAACATCAGGACGTAGCGCGACTTATTGCAGAATCATTCGCGCAAAGAGATAAACCAGAGATCGAGTTCCTCGTCGTGGAAGCAAAAAAGTCGAACTTGGAACCGAAGCCGACAGCTTTAATATTTCTGGAGGCCTAA
Protein
MSRSFDMKEEGNALIDDRGGIRRHQSMQRLQHGDQQTDESRAARVVSDHHGNLHITVKKTKPILGIAIEGGANTKHPLPRIINIHEHGAAYEAGGLEVGQLILAVDGHRVEGMQHQDVARLIAESFAQRDKPEIEFLVVEAKKSNLEPKPTALIFLEA
Summary
Uniprot
A0A0N0PC23
A0A194PMZ4
H9J0H6
A0A2A4JGI7
A0A212F704
A0A2H1WXP4
+ More
A0A1B0GNZ2 A0A0L0C3L3 A0A1Y1K1C3 A0A1B0G210 A0A139WKZ3 A0A1A9ZMV8 B0X0R6 Q17MX1 A0A0J9RW62 B4J3D1 A0A1W4U7A5 A0A2J7PDZ9 A0A1W4UIE7 A0A1W4UK57 A0A1W4U6P1 A0A1W4UIB6 A0A1B6M3N8 A0A1I8NJF2 A0A0J9UKN9 B7Z048 A0A1W4U6N5 A0A1W4UK52 A0A1W4UID3 A0A1W4U796 A0A0J9RV64 A0A0J9UKP3 U5EDC1 A0A0J9RUN6 M9PFJ0 Q9VU98 A0A0J9RUW8 A0A0R3P3X9 A0A1A9VMP5 A0A1W4WYG5 B4KYV7 M9PF56 A0A0R1DXL9 A0A1I8QAK6 W8AIZ5 W8B777 B4LDL8 A0A0P9AKR2 A0A0Q9X1S5 A0A182J8I0 A0A2M4CG24 Q7PMI9 A0A067R1S1 E0VND8 N6T5P9 A0A1Y1K428 A0A3B0JSX1 A0A151HZG3 A0A151WZZ6 E9J389 A0A151IL97 K7IQT4 A0A310SIE0 A0A151J8A0 A0A182GTZ0 A0A088A1F7 T1DRZ7 A0A182Q5Y3 A0A0L7R320 A0A182KV88 A0A0N0BHL2 A0A182I5T1 A0A182V8Q4 E2C844 A0A084WUH7 W5JEL6 A0A182FVX3 A0A182X0J5 A0A182LUM4 A0A182REK3 A0A182NV62 A0A182YSR4 A0A182KFP6 A0A182P9U7 A0A182W9L1 A0A182T2W8 A0A026X3P9 A0A1B6E6E7 U4TT61 A0A336LQM3 A0A195FUZ2 V9IDY8 A0A0J7K5G6 E2AQT2 A0A023F5U3 V9IBM0 A0A1A9Y9U1 A0A1B0BKA9 A0A1A9WCS2 A0A0A9Z0Z5
A0A1B0GNZ2 A0A0L0C3L3 A0A1Y1K1C3 A0A1B0G210 A0A139WKZ3 A0A1A9ZMV8 B0X0R6 Q17MX1 A0A0J9RW62 B4J3D1 A0A1W4U7A5 A0A2J7PDZ9 A0A1W4UIE7 A0A1W4UK57 A0A1W4U6P1 A0A1W4UIB6 A0A1B6M3N8 A0A1I8NJF2 A0A0J9UKN9 B7Z048 A0A1W4U6N5 A0A1W4UK52 A0A1W4UID3 A0A1W4U796 A0A0J9RV64 A0A0J9UKP3 U5EDC1 A0A0J9RUN6 M9PFJ0 Q9VU98 A0A0J9RUW8 A0A0R3P3X9 A0A1A9VMP5 A0A1W4WYG5 B4KYV7 M9PF56 A0A0R1DXL9 A0A1I8QAK6 W8AIZ5 W8B777 B4LDL8 A0A0P9AKR2 A0A0Q9X1S5 A0A182J8I0 A0A2M4CG24 Q7PMI9 A0A067R1S1 E0VND8 N6T5P9 A0A1Y1K428 A0A3B0JSX1 A0A151HZG3 A0A151WZZ6 E9J389 A0A151IL97 K7IQT4 A0A310SIE0 A0A151J8A0 A0A182GTZ0 A0A088A1F7 T1DRZ7 A0A182Q5Y3 A0A0L7R320 A0A182KV88 A0A0N0BHL2 A0A182I5T1 A0A182V8Q4 E2C844 A0A084WUH7 W5JEL6 A0A182FVX3 A0A182X0J5 A0A182LUM4 A0A182REK3 A0A182NV62 A0A182YSR4 A0A182KFP6 A0A182P9U7 A0A182W9L1 A0A182T2W8 A0A026X3P9 A0A1B6E6E7 U4TT61 A0A336LQM3 A0A195FUZ2 V9IDY8 A0A0J7K5G6 E2AQT2 A0A023F5U3 V9IBM0 A0A1A9Y9U1 A0A1B0BKA9 A0A1A9WCS2 A0A0A9Z0Z5
Pubmed
26354079
19121390
22118469
26108605
28004739
18362917
+ More
19820115 17510324 22936249 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17550304 24495485 12364791 24845553 20566863 23537049 21282665 20075255 26483478 20966253 20798317 24438588 20920257 23761445 25244985 24508170 25474469 25401762
19820115 17510324 22936249 17994087 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15632085 17550304 24495485 12364791 24845553 20566863 23537049 21282665 20075255 26483478 20966253 20798317 24438588 20920257 23761445 25244985 24508170 25474469 25401762
EMBL
KQ460709
KPJ12720.1
KQ459598
KPI94687.1
BABH01010409
NWSH01001661
+ More
PCG70502.1 AGBW02009947 OWR49521.1 ODYU01011429 SOQ57164.1 AJVK01014763 JRES01000945 KNC26933.1 GEZM01095944 JAV55243.1 CCAG010020724 CCAG010020725 KQ971326 KYB28491.1 DS232244 EDS38308.1 CH477203 EAT48031.1 CM002912 KMY99469.1 CH916366 EDV97230.1 NEVH01026120 PNF14561.1 GEBQ01009473 JAT30504.1 KMY99470.1 AE014296 ACL83302.1 KMY99477.1 KMY99475.1 GANO01004828 JAB55043.1 KMY99471.1 AGB94504.1 AAF49791.3 KMY99478.1 CH379069 KRT07874.1 CH933809 EDW18849.2 AGB94503.1 CM000159 KRK01835.1 GAMC01020868 JAB85687.1 GAMC01020871 JAB85684.1 CH940647 EDW69979.2 CH902618 KPU78457.1 CH964095 KRF99014.1 GGFL01000007 MBW64185.1 AAAB01008979 EAA13766.3 KK852770 KDR16921.1 DS235336 EEB14894.1 APGK01051366 APGK01051367 KB741190 ENN72998.1 GEZM01095946 JAV55241.1 OUUW01000002 SPP76456.1 KQ976699 KYM77431.1 KQ982635 KYQ53337.1 GL768059 EFZ12707.1 KQ977121 KYN05527.1 KQ760945 OAD58594.1 KQ979548 KYN21083.1 JXUM01018380 KQ560511 KXJ82051.1 GAMD01000410 JAB01181.1 AXCN02001807 KQ414664 KOC65171.1 KQ435748 KOX76431.1 APCN01003496 GL453538 EFN75897.1 ATLV01027085 KE525423 KFB53871.1 ADMH02001614 ETN61788.1 AXCM01016381 KK107019 EZA62708.1 GEDC01003829 JAS33469.1 KB630568 KB630894 ERL83686.1 ERL83932.1 UFQT01000117 SSX20374.1 KQ981241 KYN44273.1 JR039096 AEY58529.1 LBMM01013608 KMQ85562.1 GL441834 EFN64194.1 GBBI01002160 JAC16552.1 JR039095 AEY58528.1 JXJN01015863 GBHO01004652 JAG38952.1
PCG70502.1 AGBW02009947 OWR49521.1 ODYU01011429 SOQ57164.1 AJVK01014763 JRES01000945 KNC26933.1 GEZM01095944 JAV55243.1 CCAG010020724 CCAG010020725 KQ971326 KYB28491.1 DS232244 EDS38308.1 CH477203 EAT48031.1 CM002912 KMY99469.1 CH916366 EDV97230.1 NEVH01026120 PNF14561.1 GEBQ01009473 JAT30504.1 KMY99470.1 AE014296 ACL83302.1 KMY99477.1 KMY99475.1 GANO01004828 JAB55043.1 KMY99471.1 AGB94504.1 AAF49791.3 KMY99478.1 CH379069 KRT07874.1 CH933809 EDW18849.2 AGB94503.1 CM000159 KRK01835.1 GAMC01020868 JAB85687.1 GAMC01020871 JAB85684.1 CH940647 EDW69979.2 CH902618 KPU78457.1 CH964095 KRF99014.1 GGFL01000007 MBW64185.1 AAAB01008979 EAA13766.3 KK852770 KDR16921.1 DS235336 EEB14894.1 APGK01051366 APGK01051367 KB741190 ENN72998.1 GEZM01095946 JAV55241.1 OUUW01000002 SPP76456.1 KQ976699 KYM77431.1 KQ982635 KYQ53337.1 GL768059 EFZ12707.1 KQ977121 KYN05527.1 KQ760945 OAD58594.1 KQ979548 KYN21083.1 JXUM01018380 KQ560511 KXJ82051.1 GAMD01000410 JAB01181.1 AXCN02001807 KQ414664 KOC65171.1 KQ435748 KOX76431.1 APCN01003496 GL453538 EFN75897.1 ATLV01027085 KE525423 KFB53871.1 ADMH02001614 ETN61788.1 AXCM01016381 KK107019 EZA62708.1 GEDC01003829 JAS33469.1 KB630568 KB630894 ERL83686.1 ERL83932.1 UFQT01000117 SSX20374.1 KQ981241 KYN44273.1 JR039096 AEY58529.1 LBMM01013608 KMQ85562.1 GL441834 EFN64194.1 GBBI01002160 JAC16552.1 JR039095 AEY58528.1 JXJN01015863 GBHO01004652 JAG38952.1
Proteomes
UP000053240
UP000053268
UP000005204
UP000218220
UP000007151
UP000092462
+ More
UP000037069 UP000092444 UP000007266 UP000092445 UP000002320 UP000008820 UP000001070 UP000192221 UP000235965 UP000095301 UP000000803 UP000001819 UP000078200 UP000192223 UP000009192 UP000002282 UP000095300 UP000008792 UP000007801 UP000007798 UP000075880 UP000007062 UP000027135 UP000009046 UP000019118 UP000268350 UP000078540 UP000075809 UP000078542 UP000002358 UP000078492 UP000069940 UP000249989 UP000005203 UP000075886 UP000053825 UP000075882 UP000053105 UP000075840 UP000075903 UP000008237 UP000030765 UP000000673 UP000069272 UP000076407 UP000075883 UP000075900 UP000075884 UP000076408 UP000075881 UP000075885 UP000075920 UP000075901 UP000053097 UP000030742 UP000078541 UP000036403 UP000000311 UP000092443 UP000092460 UP000091820
UP000037069 UP000092444 UP000007266 UP000092445 UP000002320 UP000008820 UP000001070 UP000192221 UP000235965 UP000095301 UP000000803 UP000001819 UP000078200 UP000192223 UP000009192 UP000002282 UP000095300 UP000008792 UP000007801 UP000007798 UP000075880 UP000007062 UP000027135 UP000009046 UP000019118 UP000268350 UP000078540 UP000075809 UP000078542 UP000002358 UP000078492 UP000069940 UP000249989 UP000005203 UP000075886 UP000053825 UP000075882 UP000053105 UP000075840 UP000075903 UP000008237 UP000030765 UP000000673 UP000069272 UP000076407 UP000075883 UP000075900 UP000075884 UP000076408 UP000075881 UP000075885 UP000075920 UP000075901 UP000053097 UP000030742 UP000078541 UP000036403 UP000000311 UP000092443 UP000092460 UP000091820
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
A0A0N0PC23
A0A194PMZ4
H9J0H6
A0A2A4JGI7
A0A212F704
A0A2H1WXP4
+ More
A0A1B0GNZ2 A0A0L0C3L3 A0A1Y1K1C3 A0A1B0G210 A0A139WKZ3 A0A1A9ZMV8 B0X0R6 Q17MX1 A0A0J9RW62 B4J3D1 A0A1W4U7A5 A0A2J7PDZ9 A0A1W4UIE7 A0A1W4UK57 A0A1W4U6P1 A0A1W4UIB6 A0A1B6M3N8 A0A1I8NJF2 A0A0J9UKN9 B7Z048 A0A1W4U6N5 A0A1W4UK52 A0A1W4UID3 A0A1W4U796 A0A0J9RV64 A0A0J9UKP3 U5EDC1 A0A0J9RUN6 M9PFJ0 Q9VU98 A0A0J9RUW8 A0A0R3P3X9 A0A1A9VMP5 A0A1W4WYG5 B4KYV7 M9PF56 A0A0R1DXL9 A0A1I8QAK6 W8AIZ5 W8B777 B4LDL8 A0A0P9AKR2 A0A0Q9X1S5 A0A182J8I0 A0A2M4CG24 Q7PMI9 A0A067R1S1 E0VND8 N6T5P9 A0A1Y1K428 A0A3B0JSX1 A0A151HZG3 A0A151WZZ6 E9J389 A0A151IL97 K7IQT4 A0A310SIE0 A0A151J8A0 A0A182GTZ0 A0A088A1F7 T1DRZ7 A0A182Q5Y3 A0A0L7R320 A0A182KV88 A0A0N0BHL2 A0A182I5T1 A0A182V8Q4 E2C844 A0A084WUH7 W5JEL6 A0A182FVX3 A0A182X0J5 A0A182LUM4 A0A182REK3 A0A182NV62 A0A182YSR4 A0A182KFP6 A0A182P9U7 A0A182W9L1 A0A182T2W8 A0A026X3P9 A0A1B6E6E7 U4TT61 A0A336LQM3 A0A195FUZ2 V9IDY8 A0A0J7K5G6 E2AQT2 A0A023F5U3 V9IBM0 A0A1A9Y9U1 A0A1B0BKA9 A0A1A9WCS2 A0A0A9Z0Z5
A0A1B0GNZ2 A0A0L0C3L3 A0A1Y1K1C3 A0A1B0G210 A0A139WKZ3 A0A1A9ZMV8 B0X0R6 Q17MX1 A0A0J9RW62 B4J3D1 A0A1W4U7A5 A0A2J7PDZ9 A0A1W4UIE7 A0A1W4UK57 A0A1W4U6P1 A0A1W4UIB6 A0A1B6M3N8 A0A1I8NJF2 A0A0J9UKN9 B7Z048 A0A1W4U6N5 A0A1W4UK52 A0A1W4UID3 A0A1W4U796 A0A0J9RV64 A0A0J9UKP3 U5EDC1 A0A0J9RUN6 M9PFJ0 Q9VU98 A0A0J9RUW8 A0A0R3P3X9 A0A1A9VMP5 A0A1W4WYG5 B4KYV7 M9PF56 A0A0R1DXL9 A0A1I8QAK6 W8AIZ5 W8B777 B4LDL8 A0A0P9AKR2 A0A0Q9X1S5 A0A182J8I0 A0A2M4CG24 Q7PMI9 A0A067R1S1 E0VND8 N6T5P9 A0A1Y1K428 A0A3B0JSX1 A0A151HZG3 A0A151WZZ6 E9J389 A0A151IL97 K7IQT4 A0A310SIE0 A0A151J8A0 A0A182GTZ0 A0A088A1F7 T1DRZ7 A0A182Q5Y3 A0A0L7R320 A0A182KV88 A0A0N0BHL2 A0A182I5T1 A0A182V8Q4 E2C844 A0A084WUH7 W5JEL6 A0A182FVX3 A0A182X0J5 A0A182LUM4 A0A182REK3 A0A182NV62 A0A182YSR4 A0A182KFP6 A0A182P9U7 A0A182W9L1 A0A182T2W8 A0A026X3P9 A0A1B6E6E7 U4TT61 A0A336LQM3 A0A195FUZ2 V9IDY8 A0A0J7K5G6 E2AQT2 A0A023F5U3 V9IBM0 A0A1A9Y9U1 A0A1B0BKA9 A0A1A9WCS2 A0A0A9Z0Z5
PDB
1UFX
E-value=1.46765e-18,
Score=222
Ontologies
GO
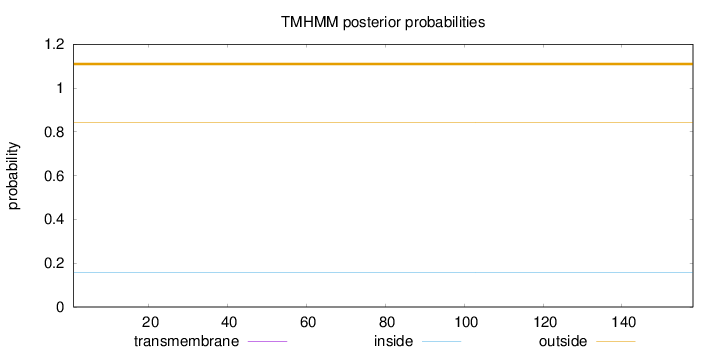
Topology
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00111
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15824
outside
1 - 158
Population Genetic Test Statistics
Pi
137.209304
Theta
147.985558
Tajima's D
-0.769197
CLR
687.441718
CSRT
0.182240887955602
Interpretation
Uncertain
