Gene
KWMTBOMO02152
Pre Gene Modal
BGIBMGA003011
Annotation
PREDICTED:_ral_guanine_nucleotide_dissociation_stimulator_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.191
Sequence
CDS
ATGAGCCAGGATGCGGAGAGCTTGCCAACATGGCGGTTGTGGGGGGAAGAGCGCGTGGACGGCGCCATCTACACCGTTTATCTGAAGAAAGTGCGTTACCATCGACCTACCAAAAGTGCTTCAAGTGAGTCTGACGACGAGATCTCACATTTAGAGTGGGAAACCGTACGTGTACGGTTCGTAAAAGCCGGCACAGTCGAGCGGCTAGTGGAGGCGCTCGCTACCGACGACGGCGAACTCGAGTCCACATACGTCAATGTGTTCCTCGCGACGTACCGGTCATTCGCGGAATGCAGTCATGTTCTGGATCTCCTGCTGCGACGGTATGAAACCCTTGCCGCTGAGGACATACTTCCCGCTGCGACTGACACATCACAACAACACCGAAAGACTCTTGTAGCGTGTCTCCACGTGTGGTTGGACACATATCCGGAGGACTTTCGGCAGCCGCCGCACCATCCCGCACTGCAACAGCTGCTCGCCTTCACGCAACTTCATTTACCCGGATCAGAATTACACTCGAAGGTATTACAGAAGCTAGCCATATTCAGTGCGGAACGAAATGGAGGTGTGCAGATGTCTGGAGGGGGCGCCGTGTGCCTTGCTCCTGGCATGCGCCTCTCCGCACATCTCACCAGCGCCTACCGCCTGCCACACGTTCCCCATCGACACTTCGCCGAACAACTTACCAGGATGGACATGGAGCTGTTCAAGAAGCTCGTGCCCCACCAGTGCCTCGGAGCCGTCTGGTCACGTAGAGATAAGGACCGTTCCCACGACGCGGCCACTGTCCTCGCAACCGTCAACCAGTTCAATGCGGTATCGTTCCGCGTCATTTCGACTGTACTCGTGGAGCTCAACTTGAAGCCGGCCGAGCGTGCCGATATTCTTGCTGCCTGGCTTGATATAGCTCGCGAACTGAGAGTTCTTAAAAACTTTTCATCGCTCAAAGCGATTATCTCAGGGTTGCAGAGCAATCCGGTTTATAGACTGAGAAAAACGTGGGCGACTTTACAAAAAGAAAAAGCTGAGCTTTACGATGAACTAGCCCGAATTTTCAGTGAAGACAATAACAGATGGGCTCAGAGGGAGTTGCTGATGCGCGAAGGTACTGCTAAATTTGCTGATACTGTGGGAGAAAACGATCGTCAATTACAAAAACTTCTTCACGAGCGCGGCTCACCAGGTGTCAGCCACGGGACCATTCCATATCTCGGCACGTTTTTGACCGATCTCACAATGATAGATACTGCCATTCCGGATACAGTAACCGACGGTTTGATTAATTTTGATAAGAAACGTAAAGAATTCGAAGTGCTGGCTCAGATCAAATTACTGCAAGGAGCTGCCAACGCCTACCATTTACCCTCCGATCCGATGTTTGACCGGTGGTTTGATTCGGTTATTGTGCTTGATGACAGGGAAGCATATCAGCTATCTTGTCAAATTGAGCCACCGACGAACCCACCCAAAGAGAGACGACCAAAGAAGATAAACGACGCCAACGTTCAAAGTCATAGGAAAAACGACTCGATCGCCAGCACGAGTTCTAGCAATAGTTCACAATTCTTTTGCGAAACCGATGGAACAACTACCGCTTGGAAGAGAGATAGTCTTGAAACGCGAATCTCTCCGACAAGGCTGTCACACGGTTCTTCGTCTTCATCGCTACCGTCTCTGGATGTCTCATTGTCCAGTGCTAACAGTGGACAGAAGCCAAACGGCAAAGCTGGGGGCAGTCCTGCACATCCGGGTGTTCGAAGTGGCCCAGACTTTTATATAATACGAGTGACATATGAGTCGGATTGTGCAGAAACCGAAGGCGTTGTGTTGTACAAGTCTATTATGTTAAGTAACAATGAACGCACGCCTCAAGTGATTCGGAACGCAATGCTCAAACTCAATCTTGATGGAGATCCCGATTCTTATACCTTGGCTCAAGTACTACTCGAACGAGAGATGTTGCTCCCATCGAATGCAAATGTCTACTACGCTGTTAATACCGCTTATAATTTGAACTTCATTTTACGGCCTCGGAAGAACCAGCAGCAAGAGCCAGTGGTTAATGGGAAGTCGAAAGGGAAACGAAACTAA
Protein
MSQDAESLPTWRLWGEERVDGAIYTVYLKKVRYHRPTKSASSESDDEISHLEWETVRVRFVKAGTVERLVEALATDDGELESTYVNVFLATYRSFAECSHVLDLLLRRYETLAAEDILPAATDTSQQHRKTLVACLHVWLDTYPEDFRQPPHHPALQQLLAFTQLHLPGSELHSKVLQKLAIFSAERNGGVQMSGGGAVCLAPGMRLSAHLTSAYRLPHVPHRHFAEQLTRMDMELFKKLVPHQCLGAVWSRRDKDRSHDAATVLATVNQFNAVSFRVISTVLVELNLKPAERADILAAWLDIARELRVLKNFSSLKAIISGLQSNPVYRLRKTWATLQKEKAELYDELARIFSEDNNRWAQRELLMREGTAKFADTVGENDRQLQKLLHERGSPGVSHGTIPYLGTFLTDLTMIDTAIPDTVTDGLINFDKKRKEFEVLAQIKLLQGAANAYHLPSDPMFDRWFDSVIVLDDREAYQLSCQIEPPTNPPKERRPKKINDANVQSHRKNDSIASTSSSNSSQFFCETDGTTTAWKRDSLETRISPTRLSHGSSSSSLPSLDVSLSSANSGQKPNGKAGGSPAHPGVRSGPDFYIIRVTYESDCAETEGVVLYKSIMLSNNERTPQVIRNAMLKLNLDGDPDSYTLAQVLLEREMLLPSNANVYYAVNTAYNLNFILRPRKNQQQEPVVNGKSKGKRN
Summary
Uniprot
H9J0H4
A0A2A4J4J4
A0A2A4J540
A0A2A4J5W3
A0A2H1VMQ2
A0A0N1I8Q5
+ More
A0A194PMN3 A0A212EJB0 A0A2A4J5T8 A0A0L7KP69 A0A1Y1LF26 A0A1B6C8Z3 A0A1B6CX98 A0A1B6F061 A0A2J7PN29 A0A2J7PN26 A0A0C9RR14 A0A0C9QXE4 A0A0C9RTT4 D6WEA5 A0A067R225 A0A151HZC5 A0A0M9A3D8 A0A158NLU0 A0A023F3B5 A0A0V0GCP7 A0A023F3J6 A0A2M4CXT2 E2C840 A0A3L8DHX4 A0A1Q3FU09 A0A1Q3FWK1 A0A1Q3FU73 A0A026X382 E2AQT6 A0A182J8D4 A0A182YIW5 A0A151WZL1 A0A151J895 A0A154PNB2 A0A084VD04 A0A151ILA0 A0A182GD60 A0A195FV23 W5JRC1 A0A182QGR8 A0A2M4CN86 A0A0A9YFA2 A0A182REJ9 A0A2M4CM18 A0A182NV68 A0A182P9V0 Q5TPG0 A0A182X0J1 A0A182UWC0 A0A0K8TIT6 A0A182KV92 A0A182U3E1 A0A182I5S7 A0A182W9K7 Q17MY4 A0A182K6A0 A0A0J7KY49 A0A2M4A831 A0A2M4A8M6 A0A182LXN5 A0A1L8DMC1 A0A310S7M3 K7IQT7 A0A0A9YLB9 A0A034VJQ0 A0A0K8UZH1 T1HX06 A0A0A1WPR3 A0A1L8DM35 A0A336MNK3 A0A1L8DMA9 A0A034VME6 A0A034VKS4 A0A0K8VC63 A0A0A1WR69 A0A2P8Z0S7 E9J394 A0A2A3EHW2 B0WG92 A0A1J1I0C4 U4TYI6 N6TZZ9 A0A0P5X2D5 A0A0P5FU45 A0A0P5HIC1 A0A0N7ZJ25 E9GFB3 A0A2S2Q327 J9JRQ4 A0A2H8TKB2 A0A2H8TY88 A0A2S2P9R5 A0A0Q5U863 A0A224X9Z1
A0A194PMN3 A0A212EJB0 A0A2A4J5T8 A0A0L7KP69 A0A1Y1LF26 A0A1B6C8Z3 A0A1B6CX98 A0A1B6F061 A0A2J7PN29 A0A2J7PN26 A0A0C9RR14 A0A0C9QXE4 A0A0C9RTT4 D6WEA5 A0A067R225 A0A151HZC5 A0A0M9A3D8 A0A158NLU0 A0A023F3B5 A0A0V0GCP7 A0A023F3J6 A0A2M4CXT2 E2C840 A0A3L8DHX4 A0A1Q3FU09 A0A1Q3FWK1 A0A1Q3FU73 A0A026X382 E2AQT6 A0A182J8D4 A0A182YIW5 A0A151WZL1 A0A151J895 A0A154PNB2 A0A084VD04 A0A151ILA0 A0A182GD60 A0A195FV23 W5JRC1 A0A182QGR8 A0A2M4CN86 A0A0A9YFA2 A0A182REJ9 A0A2M4CM18 A0A182NV68 A0A182P9V0 Q5TPG0 A0A182X0J1 A0A182UWC0 A0A0K8TIT6 A0A182KV92 A0A182U3E1 A0A182I5S7 A0A182W9K7 Q17MY4 A0A182K6A0 A0A0J7KY49 A0A2M4A831 A0A2M4A8M6 A0A182LXN5 A0A1L8DMC1 A0A310S7M3 K7IQT7 A0A0A9YLB9 A0A034VJQ0 A0A0K8UZH1 T1HX06 A0A0A1WPR3 A0A1L8DM35 A0A336MNK3 A0A1L8DMA9 A0A034VME6 A0A034VKS4 A0A0K8VC63 A0A0A1WR69 A0A2P8Z0S7 E9J394 A0A2A3EHW2 B0WG92 A0A1J1I0C4 U4TYI6 N6TZZ9 A0A0P5X2D5 A0A0P5FU45 A0A0P5HIC1 A0A0N7ZJ25 E9GFB3 A0A2S2Q327 J9JRQ4 A0A2H8TKB2 A0A2H8TY88 A0A2S2P9R5 A0A0Q5U863 A0A224X9Z1
Pubmed
EMBL
BABH01010415
NWSH01003201
PCG66799.1
PCG66798.1
PCG66802.1
ODYU01003388
+ More
SOQ42088.1 KQ460709 KPJ12718.1 KQ459598 KPI94691.1 AGBW02014507 OWR41548.1 PCG66800.1 JTDY01008089 KOB64749.1 GEZM01059416 JAV71488.1 GEDC01027346 JAS09952.1 GEDC01019234 GEDC01011640 GEDC01005825 GEDC01004469 JAS18064.1 JAS25658.1 JAS31473.1 JAS32829.1 GECZ01026214 JAS43555.1 NEVH01023958 PNF17742.1 PNF17743.1 GBYB01010980 GBYB01010984 JAG80747.1 JAG80751.1 GBYB01005327 JAG75094.1 GBYB01010981 JAG80748.1 KQ971326 EFA00851.2 KK852770 KDR16917.1 KQ976699 KYM77426.1 KQ435748 KOX76425.1 ADTU01019847 ADTU01019848 GBBI01002802 JAC15910.1 GECL01000983 JAP05141.1 GBBI01002801 JAC15911.1 GGFL01005965 MBW70143.1 GL453538 EFN75893.1 QOIP01000007 RLU20017.1 GFDL01003956 JAV31089.1 GFDL01003199 JAV31846.1 GFDL01003957 JAV31088.1 KK107019 EZA62712.1 GL441834 EFN64198.1 KQ982635 KYQ53333.1 KQ979548 KYN21077.1 KQ434978 KZC12954.1 ATLV01011092 ATLV01011093 ATLV01011094 KE524639 KFB35848.1 KQ977121 KYN05524.1 JXUM01055230 JXUM01055231 KQ561857 KXJ77307.1 KQ981241 KYN44276.1 ADMH02000692 ETN65299.1 AXCN02001805 GGFL01002624 MBW66802.1 GBHO01013273 GBRD01000378 GDHC01006903 JAG30331.1 JAG65443.1 JAQ11726.1 GGFL01002199 MBW66377.1 AAAB01008979 EAL39337.3 GBRD01000377 JAG65444.1 APCN01003495 CH477203 EAT48018.1 LBMM01001960 KMQ95472.1 GGFK01003569 MBW36890.1 GGFK01003798 MBW37119.1 AXCM01009161 GFDF01006476 JAV07608.1 KQ765234 OAD53999.1 GBHO01013274 JAG30330.1 GAKP01016268 JAC42684.1 GDHF01020313 JAI32001.1 ACPB03001559 GBXI01013248 GBXI01007512 GBXI01004281 JAD01044.1 JAD06780.1 JAD10011.1 GFDF01006613 JAV07471.1 UFQT01001862 SSX32034.1 GFDF01006614 JAV07470.1 GAKP01016269 JAC42683.1 GAKP01016270 JAC42682.1 GDHF01015765 JAI36549.1 GBXI01013272 JAD01020.1 PYGN01000252 PSN50080.1 GL768059 EFZ12713.1 KZ288250 PBC31064.1 DS231923 EDS26788.1 CVRI01000037 CRK93643.1 KB631657 ERL85088.1 APGK01045058 APGK01045059 KB741028 ENN74870.1 GDIP01078840 JAM24875.1 GDIP01143630 GDIP01119128 LRGB01002076 JAJ79772.1 KZS09338.1 GDIQ01227588 GDIQ01184452 JAK24137.1 GDIP01240537 JAI82864.1 GL732542 EFX81840.1 GGMS01002847 MBY72050.1 ABLF02038419 ABLF02038428 GFXV01002792 MBW14597.1 GFXV01007468 MBW19273.1 GGMR01013568 MBY26187.1 CH954178 KQS43924.1 GFTR01007281 JAW09145.1
SOQ42088.1 KQ460709 KPJ12718.1 KQ459598 KPI94691.1 AGBW02014507 OWR41548.1 PCG66800.1 JTDY01008089 KOB64749.1 GEZM01059416 JAV71488.1 GEDC01027346 JAS09952.1 GEDC01019234 GEDC01011640 GEDC01005825 GEDC01004469 JAS18064.1 JAS25658.1 JAS31473.1 JAS32829.1 GECZ01026214 JAS43555.1 NEVH01023958 PNF17742.1 PNF17743.1 GBYB01010980 GBYB01010984 JAG80747.1 JAG80751.1 GBYB01005327 JAG75094.1 GBYB01010981 JAG80748.1 KQ971326 EFA00851.2 KK852770 KDR16917.1 KQ976699 KYM77426.1 KQ435748 KOX76425.1 ADTU01019847 ADTU01019848 GBBI01002802 JAC15910.1 GECL01000983 JAP05141.1 GBBI01002801 JAC15911.1 GGFL01005965 MBW70143.1 GL453538 EFN75893.1 QOIP01000007 RLU20017.1 GFDL01003956 JAV31089.1 GFDL01003199 JAV31846.1 GFDL01003957 JAV31088.1 KK107019 EZA62712.1 GL441834 EFN64198.1 KQ982635 KYQ53333.1 KQ979548 KYN21077.1 KQ434978 KZC12954.1 ATLV01011092 ATLV01011093 ATLV01011094 KE524639 KFB35848.1 KQ977121 KYN05524.1 JXUM01055230 JXUM01055231 KQ561857 KXJ77307.1 KQ981241 KYN44276.1 ADMH02000692 ETN65299.1 AXCN02001805 GGFL01002624 MBW66802.1 GBHO01013273 GBRD01000378 GDHC01006903 JAG30331.1 JAG65443.1 JAQ11726.1 GGFL01002199 MBW66377.1 AAAB01008979 EAL39337.3 GBRD01000377 JAG65444.1 APCN01003495 CH477203 EAT48018.1 LBMM01001960 KMQ95472.1 GGFK01003569 MBW36890.1 GGFK01003798 MBW37119.1 AXCM01009161 GFDF01006476 JAV07608.1 KQ765234 OAD53999.1 GBHO01013274 JAG30330.1 GAKP01016268 JAC42684.1 GDHF01020313 JAI32001.1 ACPB03001559 GBXI01013248 GBXI01007512 GBXI01004281 JAD01044.1 JAD06780.1 JAD10011.1 GFDF01006613 JAV07471.1 UFQT01001862 SSX32034.1 GFDF01006614 JAV07470.1 GAKP01016269 JAC42683.1 GAKP01016270 JAC42682.1 GDHF01015765 JAI36549.1 GBXI01013272 JAD01020.1 PYGN01000252 PSN50080.1 GL768059 EFZ12713.1 KZ288250 PBC31064.1 DS231923 EDS26788.1 CVRI01000037 CRK93643.1 KB631657 ERL85088.1 APGK01045058 APGK01045059 KB741028 ENN74870.1 GDIP01078840 JAM24875.1 GDIP01143630 GDIP01119128 LRGB01002076 JAJ79772.1 KZS09338.1 GDIQ01227588 GDIQ01184452 JAK24137.1 GDIP01240537 JAI82864.1 GL732542 EFX81840.1 GGMS01002847 MBY72050.1 ABLF02038419 ABLF02038428 GFXV01002792 MBW14597.1 GFXV01007468 MBW19273.1 GGMR01013568 MBY26187.1 CH954178 KQS43924.1 GFTR01007281 JAW09145.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000235965 UP000007266 UP000027135 UP000078540 UP000053105 UP000005205 UP000008237 UP000279307 UP000053097 UP000000311 UP000075880 UP000076408 UP000075809 UP000078492 UP000076502 UP000030765 UP000078542 UP000069940 UP000249989 UP000078541 UP000000673 UP000075886 UP000075900 UP000075884 UP000075885 UP000007062 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000075920 UP000008820 UP000075881 UP000036403 UP000075883 UP000002358 UP000015103 UP000245037 UP000242457 UP000002320 UP000183832 UP000030742 UP000019118 UP000076858 UP000000305 UP000007819 UP000008711
UP000235965 UP000007266 UP000027135 UP000078540 UP000053105 UP000005205 UP000008237 UP000279307 UP000053097 UP000000311 UP000075880 UP000076408 UP000075809 UP000078492 UP000076502 UP000030765 UP000078542 UP000069940 UP000249989 UP000078541 UP000000673 UP000075886 UP000075900 UP000075884 UP000075885 UP000007062 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000075920 UP000008820 UP000075881 UP000036403 UP000075883 UP000002358 UP000015103 UP000245037 UP000242457 UP000002320 UP000183832 UP000030742 UP000019118 UP000076858 UP000000305 UP000007819 UP000008711
PRIDE
Interpro
IPR023578
Ras_GEF_dom_sf
+ More
IPR036964 RASGEF_cat_dom_sf
IPR008937 Ras-like_GEF
IPR001895 RASGEF_cat_dom
IPR000651 Ras-like_Gua-exchang_fac_N
IPR000159 RA_dom
IPR019804 Ras_G-nucl-exch_fac_CS
IPR029071 Ubiquitin-like_domsf
IPR003124 WH2_dom
IPR001478 PDZ
IPR036034 PDZ_sf
IPR018247 EF_Hand_1_Ca_BS
IPR036964 RASGEF_cat_dom_sf
IPR008937 Ras-like_GEF
IPR001895 RASGEF_cat_dom
IPR000651 Ras-like_Gua-exchang_fac_N
IPR000159 RA_dom
IPR019804 Ras_G-nucl-exch_fac_CS
IPR029071 Ubiquitin-like_domsf
IPR003124 WH2_dom
IPR001478 PDZ
IPR036034 PDZ_sf
IPR018247 EF_Hand_1_Ca_BS
Gene 3D
ProteinModelPortal
H9J0H4
A0A2A4J4J4
A0A2A4J540
A0A2A4J5W3
A0A2H1VMQ2
A0A0N1I8Q5
+ More
A0A194PMN3 A0A212EJB0 A0A2A4J5T8 A0A0L7KP69 A0A1Y1LF26 A0A1B6C8Z3 A0A1B6CX98 A0A1B6F061 A0A2J7PN29 A0A2J7PN26 A0A0C9RR14 A0A0C9QXE4 A0A0C9RTT4 D6WEA5 A0A067R225 A0A151HZC5 A0A0M9A3D8 A0A158NLU0 A0A023F3B5 A0A0V0GCP7 A0A023F3J6 A0A2M4CXT2 E2C840 A0A3L8DHX4 A0A1Q3FU09 A0A1Q3FWK1 A0A1Q3FU73 A0A026X382 E2AQT6 A0A182J8D4 A0A182YIW5 A0A151WZL1 A0A151J895 A0A154PNB2 A0A084VD04 A0A151ILA0 A0A182GD60 A0A195FV23 W5JRC1 A0A182QGR8 A0A2M4CN86 A0A0A9YFA2 A0A182REJ9 A0A2M4CM18 A0A182NV68 A0A182P9V0 Q5TPG0 A0A182X0J1 A0A182UWC0 A0A0K8TIT6 A0A182KV92 A0A182U3E1 A0A182I5S7 A0A182W9K7 Q17MY4 A0A182K6A0 A0A0J7KY49 A0A2M4A831 A0A2M4A8M6 A0A182LXN5 A0A1L8DMC1 A0A310S7M3 K7IQT7 A0A0A9YLB9 A0A034VJQ0 A0A0K8UZH1 T1HX06 A0A0A1WPR3 A0A1L8DM35 A0A336MNK3 A0A1L8DMA9 A0A034VME6 A0A034VKS4 A0A0K8VC63 A0A0A1WR69 A0A2P8Z0S7 E9J394 A0A2A3EHW2 B0WG92 A0A1J1I0C4 U4TYI6 N6TZZ9 A0A0P5X2D5 A0A0P5FU45 A0A0P5HIC1 A0A0N7ZJ25 E9GFB3 A0A2S2Q327 J9JRQ4 A0A2H8TKB2 A0A2H8TY88 A0A2S2P9R5 A0A0Q5U863 A0A224X9Z1
A0A194PMN3 A0A212EJB0 A0A2A4J5T8 A0A0L7KP69 A0A1Y1LF26 A0A1B6C8Z3 A0A1B6CX98 A0A1B6F061 A0A2J7PN29 A0A2J7PN26 A0A0C9RR14 A0A0C9QXE4 A0A0C9RTT4 D6WEA5 A0A067R225 A0A151HZC5 A0A0M9A3D8 A0A158NLU0 A0A023F3B5 A0A0V0GCP7 A0A023F3J6 A0A2M4CXT2 E2C840 A0A3L8DHX4 A0A1Q3FU09 A0A1Q3FWK1 A0A1Q3FU73 A0A026X382 E2AQT6 A0A182J8D4 A0A182YIW5 A0A151WZL1 A0A151J895 A0A154PNB2 A0A084VD04 A0A151ILA0 A0A182GD60 A0A195FV23 W5JRC1 A0A182QGR8 A0A2M4CN86 A0A0A9YFA2 A0A182REJ9 A0A2M4CM18 A0A182NV68 A0A182P9V0 Q5TPG0 A0A182X0J1 A0A182UWC0 A0A0K8TIT6 A0A182KV92 A0A182U3E1 A0A182I5S7 A0A182W9K7 Q17MY4 A0A182K6A0 A0A0J7KY49 A0A2M4A831 A0A2M4A8M6 A0A182LXN5 A0A1L8DMC1 A0A310S7M3 K7IQT7 A0A0A9YLB9 A0A034VJQ0 A0A0K8UZH1 T1HX06 A0A0A1WPR3 A0A1L8DM35 A0A336MNK3 A0A1L8DMA9 A0A034VME6 A0A034VKS4 A0A0K8VC63 A0A0A1WR69 A0A2P8Z0S7 E9J394 A0A2A3EHW2 B0WG92 A0A1J1I0C4 U4TYI6 N6TZZ9 A0A0P5X2D5 A0A0P5FU45 A0A0P5HIC1 A0A0N7ZJ25 E9GFB3 A0A2S2Q327 J9JRQ4 A0A2H8TKB2 A0A2H8TY88 A0A2S2P9R5 A0A0Q5U863 A0A224X9Z1
PDB
5CM9
E-value=1.43903e-53,
Score=532
Ontologies
GO
PANTHER
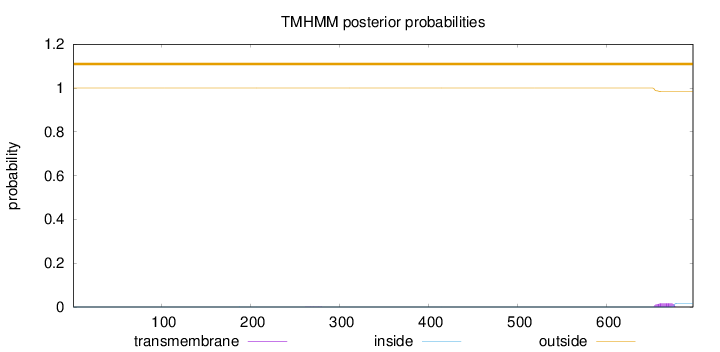
Topology
Length:
697
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35499
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00037
outside
1 - 697
Population Genetic Test Statistics
Pi
27.732514
Theta
199.053524
Tajima's D
1.300214
CLR
0.200718
CSRT
0.737513124343783
Interpretation
Uncertain
