Gene
KWMTBOMO02150
Pre Gene Modal
BGIBMGA003010
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_ATPase_WRNIP1-like_[Bombyx_mori]
Full name
ATPase WRNIP1
Alternative Name
Werner helicase-interacting protein 1
Location in the cell
Nuclear Reliability : 2.717
Sequence
CDS
ATGAGTGATTGTACAAAGACTGTGAATTGTCCTATATGTAATAGAAGTTTTGAAAATACCGAAATTGAGGAACATGTAAACAAATGTCTTTTTCTCAATTCTTGTGAAAAATCTAATTCGAAAAGACAAGGGCAACAACTATTATCTCCAAATGAAAAAAGAAAGAAAGTTGAAAAGTTAACAGCTTTGCCTCCAAACTTGAAGAGTTCACAATCACAAAAAATAAGCACAAATGAAAACATAAAAATACCTCTATCAGAGTCTTTAAGGCCAAAATCTCTGGACCAGATTGTTGGACAAAAGCAATCATTTGGCACAGATTCAATGCTCCGATCGATGCTTGAACAAAACAAGATTCCTAACATGATTTTGTGGGGTCCACCAGGCTGCGGAAAGACGTCATTAGCAAATGTTATCTCTAATATTTGTAAAAAACAAAATAATTTAAAATTTGTAAAACTATCAGCTACTACATCTGGCATCAATGATGTAAAGGAAATTGTTAAAATAGCAAAAAATGATGCACAATTTAAAAACCAGACTATTTTATTCATGGATGAAATCCACAGGTTTAACAAACTTCAACAGGACACTTTCCTGCCCCATGTTGAGAATGGTACAATAACTCTGATTGGAGCCACAACTGAAAATCCTTCTTTCAGTCTCAATAATGCTCTACTTAGTAGATGCCGGGTTGTTGTCATGGACAAACTGACCACTGAAGATGTGATGGTTATTTTAAAGAAAGCCGTTGAATACAATGAAGCAAAACTTATTAATACAAAGGAAACTGTGAACAAGAGTGATGAACCTGATCAAACCATACCCAGGTATTTAATTGAAGAAGCTTCTTTGCAATGGCTGGCAGAAATGTGTGATGGAGACGCTAGAGTCGCTTTAAGCGCCCTTGAACTGGCGCTTAATGCTCGAGATCCGGGCTCTGATGTGCACCTGAACGGCCCAGCAGTGTTAACACATGATGATATCAAAGATGGAATAAAGCGCTCGCACGTGGTATACGACAAGCACGGCGAAGAACATTACAACACGATATCTGCTATACAAAAGTGTATCCGCGCTGGAGACGATAACGCAGCGCTTTATTGGACAGCCCGAGCGATGCACGGTGGCGAAGATCCATTATATATCGCGCGACGATTTGTTAGAATGGCCTGTGAAGACATAGGCTTAGCCGATCAATCCGCATTAGTAGAAGCAGTTGCGTGTTTTCAAGGTTGCCACCAAATAGGGATGCCCGAGTGTGACGTCCTGTTGGCTCAGTGTGCTGTGAGATTAGCCCGGGCTCCGAAGAGTCGTGAAGTATACAACGCACTGAAACGGTGTCAACAATCTCTAAAAGAAACTAAAGGTCCTCTCCCACCAGTTCCATTACATTTACGGAATGCACCGACCAAGCTAATGAAGCAAATAGGGTATGGAAGTGGATACAATCTCCTGAGTAAGGAAGAATCTGGACTTACCTATTTGCCAGCAGGAGTTGAAGGTGATTATTTTCAGAAGGAATACGAAGGCGATTAA
Protein
MSDCTKTVNCPICNRSFENTEIEEHVNKCLFLNSCEKSNSKRQGQQLLSPNEKRKKVEKLTALPPNLKSSQSQKISTNENIKIPLSESLRPKSLDQIVGQKQSFGTDSMLRSMLEQNKIPNMILWGPPGCGKTSLANVISNICKKQNNLKFVKLSATTSGINDVKEIVKIAKNDAQFKNQTILFMDEIHRFNKLQQDTFLPHVENGTITLIGATTENPSFSLNNALLSRCRVVVMDKLTTEDVMVILKKAVEYNEAKLINTKETVNKSDEPDQTIPRYLIEEASLQWLAEMCDGDARVALSALELALNARDPGSDVHLNGPAVLTHDDIKDGIKRSHVVYDKHGEEHYNTISAIQKCIRAGDDNAALYWTARAMHGGEDPLYIARRFVRMACEDIGLADQSALVEAVACFQGCHQIGMPECDVLLAQCAVRLARAPKSREVYNALKRCQQSLKETKGPLPPVPLHLRNAPTKLMKQIGYGSGYNLLSKEESGLTYLPAGVEGDYFQKEYEGD
Summary
Description
Functions as a modulator of initiation or reinitiation events during DNA polymerase delta-mediated DNA synthesis. In the presence of ATP, stimulation of DNA polymerase delta-mediated DNA synthesis is decreased. Plays also a role in the innate immune defense against viruses. Stabilizes the RIG-I/DDX58 dsRNA interaction and promotes RIG-I/DDX58 'Lys-63'-linked polyubiquitination. In turn, RIG-I/DDX58 transmits the signal through mitochondrial MAVS.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Forms homooligomers (PubMed:18842586), possibly octamers. Directly interacts with POLD1, POLD2 and POLD4 (PubMed:15670210). Interacts with the N-terminal domain of WRN (By similarity). Interacts (via UBZ-type zinc finger) with monoubiquitin and polyubiquitin (PubMed:17550899, PubMed:18842586). Interacts with TRIM14 and PPP6C; these interactions positively regulate the RIG-I/DDX58 signaling pathway (PubMed:29053956).
Similarity
Belongs to the AAA ATPase family. RarA/MGS1/WRNIP1 subfamily.
Keywords
3D-structure
Acetylation
Alternative splicing
ATP-binding
Complete proteome
Cytoplasm
DNA damage
DNA replication
Hydrolase
Immunity
Innate immunity
Isopeptide bond
Metal-binding
Nucleotide-binding
Nucleus
Phosphoprotein
Reference proteome
Ubl conjugation
Zinc
Zinc-finger
Feature
chain ATPase WRNIP1
splice variant In isoform 4.
splice variant In isoform 4.
Uniprot
A0A2A4J5J3
A0A194PU82
A0A212EJ75
A0A0N1PGE5
A0A2J7Q1L6
A0A2J7Q1N3
+ More
A0A067RT66 A0A069DV19 A0A023EX55 A0A336LTW2 A0A2J7Q1J9 Q17P00 A0A0V0G9H6 A0A0J7LAV4 T1I0G0 E2A851 A0A1Q3G0L8 A0A0A9YYF7 A0A0A9Z634 A0A0K8TFQ0 A0A146LGE5 A0A151J2D3 A0A146LMN1 A0A1B6EPV5 A0A158NZU8 A0A151XBC0 A0A195BLC6 F4WIC3 A0A0M8ZVW2 A0A310SB61 A0A026WNM5 A0A195F2N3 A0A3L8DVG0 A0A195CM05 A0A088ATB3 A0A0L7R9N9 A0A2A3EIC3 A0A1B6C8Q5 A0A182WEJ5 A0A182PEP9 A0A084VIC1 A0A182R6Z5 A0A0P4VIC6 A0A0L7L436 W5J246 E2B7B1 A0A182LUJ1 A0A154NYY2 A0A182IM42 A0A182Q8F0 A0A1L8DWP1 A0A1B0CRN8 A0A182UNP0 A0A182HN02 A0A182YJB7 A0A182TZE0 A0A182WXA1 A0A182K8M6 A0A182L4A4 A0A182N2G0 A0A1Y1KPP1 A0A1S4HD89 A0A151NFA4 H0YV62 A7S6Y1 A0A218UPL3 K7G8Z8 A0A091EZK2 A0A3B3R6N0 A0A093Q4Q1 F1P1N3 C3YGI0 A0A3B3R5Y0 A0A226NJ76 A0A093GKV7 G3WPE2 A0A2K5HXZ3 A0A3B4DU16 A0A2J8WMC9 A0A2I0UE67 U3J1H6 A0A3P9AEU5 G7P4B5 A0A3L8SMJ5 G1SN62 Q96S55-3 G3RVZ9 A0A2J8MU33 A0A1V4JWG7 F7DJS2 M4APJ7 F1MGL0 A0A2K6TAU4 A0A087YBU5 A0A3M0KTS3 Q6NW46 T1JL37 A0A3B3XWU7
A0A067RT66 A0A069DV19 A0A023EX55 A0A336LTW2 A0A2J7Q1J9 Q17P00 A0A0V0G9H6 A0A0J7LAV4 T1I0G0 E2A851 A0A1Q3G0L8 A0A0A9YYF7 A0A0A9Z634 A0A0K8TFQ0 A0A146LGE5 A0A151J2D3 A0A146LMN1 A0A1B6EPV5 A0A158NZU8 A0A151XBC0 A0A195BLC6 F4WIC3 A0A0M8ZVW2 A0A310SB61 A0A026WNM5 A0A195F2N3 A0A3L8DVG0 A0A195CM05 A0A088ATB3 A0A0L7R9N9 A0A2A3EIC3 A0A1B6C8Q5 A0A182WEJ5 A0A182PEP9 A0A084VIC1 A0A182R6Z5 A0A0P4VIC6 A0A0L7L436 W5J246 E2B7B1 A0A182LUJ1 A0A154NYY2 A0A182IM42 A0A182Q8F0 A0A1L8DWP1 A0A1B0CRN8 A0A182UNP0 A0A182HN02 A0A182YJB7 A0A182TZE0 A0A182WXA1 A0A182K8M6 A0A182L4A4 A0A182N2G0 A0A1Y1KPP1 A0A1S4HD89 A0A151NFA4 H0YV62 A7S6Y1 A0A218UPL3 K7G8Z8 A0A091EZK2 A0A3B3R6N0 A0A093Q4Q1 F1P1N3 C3YGI0 A0A3B3R5Y0 A0A226NJ76 A0A093GKV7 G3WPE2 A0A2K5HXZ3 A0A3B4DU16 A0A2J8WMC9 A0A2I0UE67 U3J1H6 A0A3P9AEU5 G7P4B5 A0A3L8SMJ5 G1SN62 Q96S55-3 G3RVZ9 A0A2J8MU33 A0A1V4JWG7 F7DJS2 M4APJ7 F1MGL0 A0A2K6TAU4 A0A087YBU5 A0A3M0KTS3 Q6NW46 T1JL37 A0A3B3XWU7
EC Number
3.6.1.3
Pubmed
26354079
22118469
24845553
26334808
25474469
17510324
+ More
20798317 25401762 26823975 21347285 21719571 24508170 30249741 24438588 27129103 26227816 20920257 23761445 25244985 20966253 28004739 12364791 22293439 20360741 17615350 17381049 29240929 15592404 18563158 21709235 23749191 25069045 22002653 30282656 21993624 11301316 14702039 14574404 15489334 15670210 15592455 17081983 17550899 18842586 18220336 18669648 19413330 19369195 19690332 19608861 20068231 21269460 21406692 23186163 24275569 29053956 28112733 27062441 22398555 17495919 23542700 19393038 23594743
20798317 25401762 26823975 21347285 21719571 24508170 30249741 24438588 27129103 26227816 20920257 23761445 25244985 20966253 28004739 12364791 22293439 20360741 17615350 17381049 29240929 15592404 18563158 21709235 23749191 25069045 22002653 30282656 21993624 11301316 14702039 14574404 15489334 15670210 15592455 17081983 17550899 18842586 18220336 18669648 19413330 19369195 19690332 19608861 20068231 21269460 21406692 23186163 24275569 29053956 28112733 27062441 22398555 17495919 23542700 19393038 23594743
EMBL
NWSH01003201
PCG66794.1
KQ459598
KPI94695.1
AGBW02014506
OWR41553.1
+ More
KQ460709 KPJ12714.1 NEVH01019395 PNF22466.1 PNF22469.1 KK852432 KDR24005.1 GBGD01001223 JAC87666.1 GBBI01004945 JAC13767.1 UFQS01000195 UFQT01000195 SSX01106.1 SSX21486.1 PNF22468.1 CH477195 EAT48398.1 GECL01001446 JAP04678.1 LBMM01000101 KMR04953.1 ACPB03008948 GL437497 EFN70363.1 GFDL01001696 JAV33349.1 GBHO01006340 JAG37264.1 GBHO01006339 JAG37265.1 GBRD01001404 JAG64417.1 GDHC01013049 JAQ05580.1 KQ980404 KYN16197.1 GDHC01010573 JAQ08056.1 GECZ01029805 JAS39964.1 ADTU01005004 KQ982330 KYQ57675.1 KQ976440 KYM86490.1 GL888175 EGI66054.1 KQ435853 KOX70673.1 KQ761797 OAD56792.1 KK107144 EZA57600.1 KQ981856 KYN34646.1 QOIP01000003 RLU24316.1 KQ977580 KYN01761.1 KQ414621 KOC67578.1 KZ288253 PBC30949.1 GEDC01027422 GEDC01005857 JAS09876.1 JAS31441.1 ATLV01013344 KE524854 KFB37715.1 GDKW01002074 JAI54521.1 JTDY01003048 KOB70248.1 ADMH02002139 ETN58327.1 GL446154 EFN88396.1 AXCM01004622 KQ434777 KZC04284.1 AXCN02001071 GFDF01003246 JAV10838.1 AJWK01025067 AJWK01025068 APCN01001809 GEZM01078986 JAV62548.1 AAAB01008986 AKHW03003201 KYO35473.1 ABQF01009080 DS469590 EDO40507.1 MUZQ01000197 OWK55548.1 AGCU01036227 AGCU01036228 AGCU01036229 AGCU01036230 AGCU01036231 KK719166 KFO61734.1 KL671847 KFW83963.1 AC157662 GG666511 EEN60701.1 MCFN01000039 OXB67408.1 KL216485 KFV69858.1 AEFK01064574 AEFK01064575 NDHI03003384 PNJ70934.1 KZ505832 PKU44347.1 ADON01092884 ADON01092885 CM001279 EHH52678.1 QUSF01000013 RLW04461.1 AB056152 AK026179 AK315471 AB209723 AK223593 AL139092 CH471087 BC018923 AF218313 AAH18923.1 BAB15383.1 BAB60709.1 BAD92960.1 BAD97313.1 CABD030042957 CABD030042958 NBAG03000244 PNI63036.1 LSYS01005643 OPJ76552.1 BC150136 AAI50137.1 AYCK01003155 AYCK01003156 QRBI01000104 RMC16071.1 CU693494 BC067729 AAH67729.1 JH431429
KQ460709 KPJ12714.1 NEVH01019395 PNF22466.1 PNF22469.1 KK852432 KDR24005.1 GBGD01001223 JAC87666.1 GBBI01004945 JAC13767.1 UFQS01000195 UFQT01000195 SSX01106.1 SSX21486.1 PNF22468.1 CH477195 EAT48398.1 GECL01001446 JAP04678.1 LBMM01000101 KMR04953.1 ACPB03008948 GL437497 EFN70363.1 GFDL01001696 JAV33349.1 GBHO01006340 JAG37264.1 GBHO01006339 JAG37265.1 GBRD01001404 JAG64417.1 GDHC01013049 JAQ05580.1 KQ980404 KYN16197.1 GDHC01010573 JAQ08056.1 GECZ01029805 JAS39964.1 ADTU01005004 KQ982330 KYQ57675.1 KQ976440 KYM86490.1 GL888175 EGI66054.1 KQ435853 KOX70673.1 KQ761797 OAD56792.1 KK107144 EZA57600.1 KQ981856 KYN34646.1 QOIP01000003 RLU24316.1 KQ977580 KYN01761.1 KQ414621 KOC67578.1 KZ288253 PBC30949.1 GEDC01027422 GEDC01005857 JAS09876.1 JAS31441.1 ATLV01013344 KE524854 KFB37715.1 GDKW01002074 JAI54521.1 JTDY01003048 KOB70248.1 ADMH02002139 ETN58327.1 GL446154 EFN88396.1 AXCM01004622 KQ434777 KZC04284.1 AXCN02001071 GFDF01003246 JAV10838.1 AJWK01025067 AJWK01025068 APCN01001809 GEZM01078986 JAV62548.1 AAAB01008986 AKHW03003201 KYO35473.1 ABQF01009080 DS469590 EDO40507.1 MUZQ01000197 OWK55548.1 AGCU01036227 AGCU01036228 AGCU01036229 AGCU01036230 AGCU01036231 KK719166 KFO61734.1 KL671847 KFW83963.1 AC157662 GG666511 EEN60701.1 MCFN01000039 OXB67408.1 KL216485 KFV69858.1 AEFK01064574 AEFK01064575 NDHI03003384 PNJ70934.1 KZ505832 PKU44347.1 ADON01092884 ADON01092885 CM001279 EHH52678.1 QUSF01000013 RLW04461.1 AB056152 AK026179 AK315471 AB209723 AK223593 AL139092 CH471087 BC018923 AF218313 AAH18923.1 BAB15383.1 BAB60709.1 BAD92960.1 BAD97313.1 CABD030042957 CABD030042958 NBAG03000244 PNI63036.1 LSYS01005643 OPJ76552.1 BC150136 AAI50137.1 AYCK01003155 AYCK01003156 QRBI01000104 RMC16071.1 CU693494 BC067729 AAH67729.1 JH431429
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000235965
UP000027135
+ More
UP000008820 UP000036403 UP000015103 UP000000311 UP000078492 UP000005205 UP000075809 UP000078540 UP000007755 UP000053105 UP000053097 UP000078541 UP000279307 UP000078542 UP000005203 UP000053825 UP000242457 UP000075920 UP000075885 UP000030765 UP000075900 UP000037510 UP000000673 UP000008237 UP000075883 UP000076502 UP000075880 UP000075886 UP000092461 UP000075903 UP000075840 UP000076408 UP000075902 UP000076407 UP000075881 UP000075882 UP000075884 UP000050525 UP000007754 UP000001593 UP000197619 UP000007267 UP000052976 UP000261540 UP000053258 UP000000539 UP000001554 UP000198323 UP000053875 UP000007648 UP000233080 UP000261440 UP000016666 UP000265140 UP000009130 UP000276834 UP000001811 UP000005640 UP000001519 UP000190648 UP000002280 UP000002852 UP000009136 UP000233220 UP000028760 UP000269221 UP000000437 UP000261480
UP000008820 UP000036403 UP000015103 UP000000311 UP000078492 UP000005205 UP000075809 UP000078540 UP000007755 UP000053105 UP000053097 UP000078541 UP000279307 UP000078542 UP000005203 UP000053825 UP000242457 UP000075920 UP000075885 UP000030765 UP000075900 UP000037510 UP000000673 UP000008237 UP000075883 UP000076502 UP000075880 UP000075886 UP000092461 UP000075903 UP000075840 UP000076408 UP000075902 UP000076407 UP000075881 UP000075882 UP000075884 UP000050525 UP000007754 UP000001593 UP000197619 UP000007267 UP000052976 UP000261540 UP000053258 UP000000539 UP000001554 UP000198323 UP000053875 UP000007648 UP000233080 UP000261440 UP000016666 UP000265140 UP000009130 UP000276834 UP000001811 UP000005640 UP000001519 UP000190648 UP000002280 UP000002852 UP000009136 UP000233220 UP000028760 UP000269221 UP000000437 UP000261480
PRIDE
Interpro
ProteinModelPortal
A0A2A4J5J3
A0A194PU82
A0A212EJ75
A0A0N1PGE5
A0A2J7Q1L6
A0A2J7Q1N3
+ More
A0A067RT66 A0A069DV19 A0A023EX55 A0A336LTW2 A0A2J7Q1J9 Q17P00 A0A0V0G9H6 A0A0J7LAV4 T1I0G0 E2A851 A0A1Q3G0L8 A0A0A9YYF7 A0A0A9Z634 A0A0K8TFQ0 A0A146LGE5 A0A151J2D3 A0A146LMN1 A0A1B6EPV5 A0A158NZU8 A0A151XBC0 A0A195BLC6 F4WIC3 A0A0M8ZVW2 A0A310SB61 A0A026WNM5 A0A195F2N3 A0A3L8DVG0 A0A195CM05 A0A088ATB3 A0A0L7R9N9 A0A2A3EIC3 A0A1B6C8Q5 A0A182WEJ5 A0A182PEP9 A0A084VIC1 A0A182R6Z5 A0A0P4VIC6 A0A0L7L436 W5J246 E2B7B1 A0A182LUJ1 A0A154NYY2 A0A182IM42 A0A182Q8F0 A0A1L8DWP1 A0A1B0CRN8 A0A182UNP0 A0A182HN02 A0A182YJB7 A0A182TZE0 A0A182WXA1 A0A182K8M6 A0A182L4A4 A0A182N2G0 A0A1Y1KPP1 A0A1S4HD89 A0A151NFA4 H0YV62 A7S6Y1 A0A218UPL3 K7G8Z8 A0A091EZK2 A0A3B3R6N0 A0A093Q4Q1 F1P1N3 C3YGI0 A0A3B3R5Y0 A0A226NJ76 A0A093GKV7 G3WPE2 A0A2K5HXZ3 A0A3B4DU16 A0A2J8WMC9 A0A2I0UE67 U3J1H6 A0A3P9AEU5 G7P4B5 A0A3L8SMJ5 G1SN62 Q96S55-3 G3RVZ9 A0A2J8MU33 A0A1V4JWG7 F7DJS2 M4APJ7 F1MGL0 A0A2K6TAU4 A0A087YBU5 A0A3M0KTS3 Q6NW46 T1JL37 A0A3B3XWU7
A0A067RT66 A0A069DV19 A0A023EX55 A0A336LTW2 A0A2J7Q1J9 Q17P00 A0A0V0G9H6 A0A0J7LAV4 T1I0G0 E2A851 A0A1Q3G0L8 A0A0A9YYF7 A0A0A9Z634 A0A0K8TFQ0 A0A146LGE5 A0A151J2D3 A0A146LMN1 A0A1B6EPV5 A0A158NZU8 A0A151XBC0 A0A195BLC6 F4WIC3 A0A0M8ZVW2 A0A310SB61 A0A026WNM5 A0A195F2N3 A0A3L8DVG0 A0A195CM05 A0A088ATB3 A0A0L7R9N9 A0A2A3EIC3 A0A1B6C8Q5 A0A182WEJ5 A0A182PEP9 A0A084VIC1 A0A182R6Z5 A0A0P4VIC6 A0A0L7L436 W5J246 E2B7B1 A0A182LUJ1 A0A154NYY2 A0A182IM42 A0A182Q8F0 A0A1L8DWP1 A0A1B0CRN8 A0A182UNP0 A0A182HN02 A0A182YJB7 A0A182TZE0 A0A182WXA1 A0A182K8M6 A0A182L4A4 A0A182N2G0 A0A1Y1KPP1 A0A1S4HD89 A0A151NFA4 H0YV62 A7S6Y1 A0A218UPL3 K7G8Z8 A0A091EZK2 A0A3B3R6N0 A0A093Q4Q1 F1P1N3 C3YGI0 A0A3B3R5Y0 A0A226NJ76 A0A093GKV7 G3WPE2 A0A2K5HXZ3 A0A3B4DU16 A0A2J8WMC9 A0A2I0UE67 U3J1H6 A0A3P9AEU5 G7P4B5 A0A3L8SMJ5 G1SN62 Q96S55-3 G3RVZ9 A0A2J8MU33 A0A1V4JWG7 F7DJS2 M4APJ7 F1MGL0 A0A2K6TAU4 A0A087YBU5 A0A3M0KTS3 Q6NW46 T1JL37 A0A3B3XWU7
PDB
3PVS
E-value=3.35818e-80,
Score=761
Ontologies
GO
GO:0005524
GO:0003677
GO:0006260
GO:0006281
GO:0009378
GO:0006310
GO:0004386
GO:0016887
GO:0030174
GO:0042802
GO:0048471
GO:0000731
GO:0000784
GO:0043142
GO:0006261
GO:0008047
GO:0006282
GO:0005634
GO:0045087
GO:0046872
GO:0016020
GO:0004298
GO:0003824
GO:0006520
GO:0008483
GO:0006418
GO:0016507
GO:0048384
Topology
Subcellular location
Nucleus
Colocalizes with WRN in granular structures in the nucleus. With evidence from 19 publications.
Cytoplasm Colocalizes with WRN in granular structures in the nucleus. With evidence from 19 publications.
Cytoplasm Colocalizes with WRN in granular structures in the nucleus. With evidence from 19 publications.
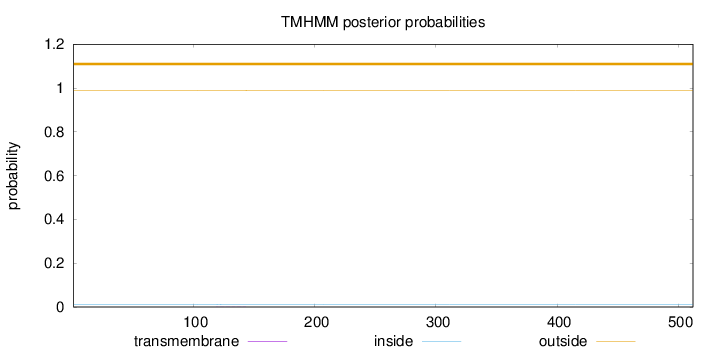
Length:
512
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01046
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01233
outside
1 - 512
Population Genetic Test Statistics
Pi
166.836152
Theta
151.778204
Tajima's D
0.352733
CLR
0.633714
CSRT
0.469776511174441
Interpretation
Uncertain
