Gene
KWMTBOMO02149
Pre Gene Modal
BGIBMGA003152
Annotation
PREDICTED:_protein_O-mannosyltransferase_1_isoform_X2_[Bombyx_mori]
Full name
Protein O-mannosyltransferase 1
Alternative Name
Dolichyl-phosphate-mannose--protein mannosyltransferase 1
Protein rotated abdomen
Protein rotated abdomen
Location in the cell
PlasmaMembrane Reliability : 4.053
Sequence
CDS
ATGGACACAGGACGGTGTGTTAAAAATCGTAAAATAAACAAGATTCGTAAGGAGGTCACAGAACCTCAGGAGTACAGTTATTCGGAGAAAACATGTAATTCCCATCGAGAAGTGGGCGATGGTGCCCCAGCTGAAAATGAAACTGAAAAACCAGAATTAATGCTGACGAACAAAATGAATCATTCTCCTTCACGCATATATTTACAGCTCAAGATTGATGTAATCGCAATAGGACTTCTAATATTGGCATTATGCACTCGTTTATATAAACTAGATGAACCTCGCCATATTGTATTTGATGAATTACATTATGGAAGATATGTTTCATTATATACTAAGGGTGTCTTTTTTTTTGATGCTCATCCACCACTTGGCAAACAGTTGTTATATCTAGCTGGTAGTATTGCTGGATATGATGGAAACTTTACTTTTGACCGGATTGGTACTCCATATACTGACAATGTTCCTGTGAAGGCTCTCAGGCTAGTTCCAGCTGTGTCAGGAAGTTTTCTAGTTTTAATTACATATTTTTTGATGCTAGAACTATCTTTACATCAATGGACTGCCATACTTGCTGCTTTATTAGTGCTCCTTGAAAATTGTTTCTTAGCACAGTCAAGGTTCATGTTGCTGGAAAGCATTCAGATACTATTTGGCTTGTGCGGTGTGTATTGTATTATTAAAAGTACAAAGAGAATTGGACTCACAGCACTTGTTTGGCTATGTGCTGCTGCTGTTTTGCTTGGTTGTTGCTTCTCGGTGAAATACTCTGGGCTGTACACATACTATTTATCATTATTCTTAGTTGGAAGACACATATGGTTGCAACTAGACCAAATAAAATCAGGAACACGTTTATTATTTTCTGCCTTGTGGCGGTTTGCGGTCTTATTGATCATCCCGCTGGCTGTGTATGTGAGCGTGTTTTACATGCATTTGTCTATGTTGCCAAAGGCTGGACCGCATGACAGTGTTATGACGAGTGCATTCCAAGCCTCATTACAGGGCGGCCTTGCGAGCATAACGCACGGACAGCCATTGCATGTCGCTCACGGCTCTCAAATAACACTCAGGCACACCCACGGCAGGACCTGTTGGCTCCATTCACACGCACACGTATATCCAGTGCGTTATGCGGACGGACGTGGCTCCTCACACCAGCAGCAAGTGACTTGTTACAGCTTCAAAGATGTCAACAATTGGTGGATTGTAAAACGGCCGGACCAAGCGTCGCTGGCCGTTTCGCGACCTCCAGACGCTATAAGACACGGAGATATCGTACAACTGTTGCATGGAATAACGAGTCGCGCTTTGAATTCCCACGATGTGGCCGCAGCTGTTTCGCCTCAATCTCAAGAAGTGTCTTGCTACATCGACTACAATGTATCGATGCCAGCTCAGAATCTTTGGCGTGTGGATATTGTGAATCGTGATTCTGAAGAGTCAACCTGGGATAGCATACGCTCGTTAGTGAGACTTGTTCACGTTGACACGGGGTCGGGCTTGCGTTACAGTGGTCGCCAACTTCCGGCTTGGGGTTTCCATCAACATGAGATTGTTGCTGACAAGGTGTTATCCCATCAAGATACAATATGGAATGTTGAAGAGCATCGCTATACCAAAGCCGATGATCGAAAAGAACGGGAACGCGAGTTAGTCACGGCTGAAATGATACCGACTGCAGTGACCCAATTAACTTTTTGGGAAAAATTTACAGAATTACAATATAAAATGATAGCACACGCACCTGATGCCCCTCACGGGCACATGTTTGCCAGCGAACCAGCTGAATGGCCTCTGCTTGTTCGTTCAATTGCATATTGGCTCTCTCCTGACTCTAATGCTCAAATACATCTGATAGGCAACATTGTAACATGGTATACTGGGACAATATCGGTAGTAGTCTACAGTGCCCTCCTGGCTATTTACTCCGTTCGAAAACGTCGAGCTTGTGAAGACTTGCCTCCATTTGTTTTGAATAAGTTTTATGAAACTGGCTATACATTATTTCTAGGTTATTGTGTTCTGAACTATGGAAATGTTGATTTAAAGGAAAATGATCTAATGAGTCTTAGGTGGAAAGATACCTGGGACTTTATATTACATAAAAAGATATAA
Protein
MDTGRCVKNRKINKIRKEVTEPQEYSYSEKTCNSHREVGDGAPAENETEKPELMLTNKMNHSPSRIYLQLKIDVIAIGLLILALCTRLYKLDEPRHIVFDELHYGRYVSLYTKGVFFFDAHPPLGKQLLYLAGSIAGYDGNFTFDRIGTPYTDNVPVKALRLVPAVSGSFLVLITYFLMLELSLHQWTAILAALLVLLENCFLAQSRFMLLESIQILFGLCGVYCIIKSTKRIGLTALVWLCAAAVLLGCCFSVKYSGLYTYYLSLFLVGRHIWLQLDQIKSGTRLLFSALWRFAVLLIIPLAVYVSVFYMHLSMLPKAGPHDSVMTSAFQASLQGGLASITHGQPLHVAHGSQITLRHTHGRTCWLHSHAHVYPVRYADGRGSSHQQQVTCYSFKDVNNWWIVKRPDQASLAVSRPPDAIRHGDIVQLLHGITSRALNSHDVAAAVSPQSQEVSCYIDYNVSMPAQNLWRVDIVNRDSEESTWDSIRSLVRLVHVDTGSGLRYSGRQLPAWGFHQHEIVADKVLSHQDTIWNVEEHRYTKADDRKERERELVTAEMIPTAVTQLTFWEKFTELQYKMIAHAPDAPHGHMFASEPAEWPLLVRSIAYWLSPDSNAQIHLIGNIVTWYTGTISVVVYSALLAIYSVRKRRACEDLPPFVLNKFYETGYTLFLGYCVLNYGNVDLKENDLMSLRWKDTWDFILHKKI
Summary
Description
Rt/POMT1 and tw/POMT2 function as a protein O-mannosyltransferase in association with each other to generate and maintain normal muscle development.
Catalytic Activity
dolichyl beta-D-mannosyl phosphate + L-seryl-[protein] = 3-O-(alpha-D-mannosyl)-L-seryl-[protein] + dolichyl phosphate + H(+)
dolichyl beta-D-mannosyl phosphate + L-threonyl-[protein] = 3-O-(alpha-D-mannosyl)-L-threonyl-[protein] + dolichyl phosphate + H(+)
dolichyl beta-D-mannosyl phosphate + L-threonyl-[protein] = 3-O-(alpha-D-mannosyl)-L-threonyl-[protein] + dolichyl phosphate + H(+)
Subunit
Interacts with tw/POMT2.
Similarity
Belongs to the glycosyltransferase 39 family.
Keywords
Complete proteome
Developmental protein
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Membrane
Reference proteome
Repeat
Transferase
Transmembrane
Transmembrane helix
Feature
chain Protein O-mannosyltransferase 1
Uniprot
A0A2H1WVF1
A0A212EJ94
A0A2A4J5U6
A0A2A3EGW3
A0A088ATG7
A0A0T6BBD0
+ More
A0A2J7Q1P0 F4WIC4 A0A151XBF5 D6WKM8 A0A158NZU9 A0A023F3C7 A0A026WRA1 A0A195CM25 E2B7B0 A0A195F241 K7INM0 A0A154NX72 A0A151J2J5 J3JUS3 N6UCU0 A0A232ELW1 U4U6M1 A0A1Y1LCU5 E0VJ18 A0A0C9Q470 A0A131XGA6 A0A1B6DQK1 A0A1B6KKG3 A0A0L0CS09 L7M7K2 A0A146KPU2 A0A224YXG8 A0A131YZ60 B4KUQ5 A0A0J9RTB0 B4QQ77 B4HEG6 A0A0A1WD25 A0A3B0JUQ1 Q9VTK2 J9JYV6 A0A1E1XKQ7 A0A067RVV3 B4GZH3 B4PF70 A0A1A9UPF5 A0A1A9WGH5 B4LCN8 V5HMB9 A0A1E1X503 B7PZ28 A0A0L7R9R8 Q2LZ62 A0A2R7VYT2 A0A1B0G158 B3NGX3 A0A0M4F0N3 V5HA07 B3M490 T1JMV6 A0A2R5L9K6 B4MKP4 W8C4P4 A0A0K8UAP7 A0A1J1I054 A0A1I8NS44 A0A1I8NS45 B4J2K3 A0A1B6EE73 A0A336MDJ3 A0A336L2C7 A0A310SJW1 A0A034VD69 A0A195BMH9 A0A182K5A9 A0A182I5S5 A0A1W4W798 A0A182REJ6 A0A182V2U2 A0A182YIW7 A0A0P4VZT5 A0A182MN00 A0A182X0I8 A0A0P4WQZ7 Q7Q222 A0A182UJ28 A0A182W9K5 A0A182S7S1 B0WG88 A0A182NV70 A0A182P9V2 A0A0P4WVR7 A0A182QJ90 T1K338 A0A1S4EX29 A0A0P5MSZ4 A0A0C9R7N7
A0A2J7Q1P0 F4WIC4 A0A151XBF5 D6WKM8 A0A158NZU9 A0A023F3C7 A0A026WRA1 A0A195CM25 E2B7B0 A0A195F241 K7INM0 A0A154NX72 A0A151J2J5 J3JUS3 N6UCU0 A0A232ELW1 U4U6M1 A0A1Y1LCU5 E0VJ18 A0A0C9Q470 A0A131XGA6 A0A1B6DQK1 A0A1B6KKG3 A0A0L0CS09 L7M7K2 A0A146KPU2 A0A224YXG8 A0A131YZ60 B4KUQ5 A0A0J9RTB0 B4QQ77 B4HEG6 A0A0A1WD25 A0A3B0JUQ1 Q9VTK2 J9JYV6 A0A1E1XKQ7 A0A067RVV3 B4GZH3 B4PF70 A0A1A9UPF5 A0A1A9WGH5 B4LCN8 V5HMB9 A0A1E1X503 B7PZ28 A0A0L7R9R8 Q2LZ62 A0A2R7VYT2 A0A1B0G158 B3NGX3 A0A0M4F0N3 V5HA07 B3M490 T1JMV6 A0A2R5L9K6 B4MKP4 W8C4P4 A0A0K8UAP7 A0A1J1I054 A0A1I8NS44 A0A1I8NS45 B4J2K3 A0A1B6EE73 A0A336MDJ3 A0A336L2C7 A0A310SJW1 A0A034VD69 A0A195BMH9 A0A182K5A9 A0A182I5S5 A0A1W4W798 A0A182REJ6 A0A182V2U2 A0A182YIW7 A0A0P4VZT5 A0A182MN00 A0A182X0I8 A0A0P4WQZ7 Q7Q222 A0A182UJ28 A0A182W9K5 A0A182S7S1 B0WG88 A0A182NV70 A0A182P9V2 A0A0P4WVR7 A0A182QJ90 T1K338 A0A1S4EX29 A0A0P5MSZ4 A0A0C9R7N7
EC Number
2.4.1.109
Pubmed
22118469
21719571
18362917
19820115
21347285
25474469
+ More
24508170 30249741 20798317 20075255 22516182 23537049 28648823 28004739 20566863 28049606 26108605 25576852 26823975 28797301 26830274 17994087 22936249 25830018 8650217 15271988 10731132 12537572 12537569 16219785 29209593 24845553 17550304 18057021 25765539 28503490 15632085 24495485 25348373 25244985 12364791 14747013 17210077
24508170 30249741 20798317 20075255 22516182 23537049 28648823 28004739 20566863 28049606 26108605 25576852 26823975 28797301 26830274 17994087 22936249 25830018 8650217 15271988 10731132 12537572 12537569 16219785 29209593 24845553 17550304 18057021 25765539 28503490 15632085 24495485 25348373 25244985 12364791 14747013 17210077
EMBL
ODYU01011353
SOQ57045.1
AGBW02014506
OWR41554.1
NWSH01003201
PCG66793.1
+ More
KZ288253 PBC30948.1 LJIG01002331 KRT84609.1 NEVH01019395 PNF22479.1 GL888175 EGI66055.1 KQ982330 KYQ57674.1 KQ971342 EFA03006.2 ADTU01005005 ADTU01005006 GBBI01002782 JAC15930.1 KK107144 QOIP01000003 EZA57599.1 RLU24315.1 KQ977580 KYN01760.1 GL446154 EFN88395.1 KQ981856 KYN34645.1 KQ434777 KZC04285.1 KQ980404 KYN16198.1 BT126988 AEE61950.1 APGK01034125 KB740904 ENN78451.1 NNAY01003486 OXU19345.1 KB632006 ERL87963.1 GEZM01059416 JAV71489.1 DS235219 EEB13374.1 GBYB01008836 GBYB01008838 JAG78603.1 JAG78605.1 GEFH01002408 JAP66173.1 GEDC01016863 GEDC01009346 JAS20435.1 JAS27952.1 GEBQ01028051 GEBQ01003489 JAT11926.1 JAT36488.1 JRES01000005 KNC34982.1 GACK01005177 JAA59857.1 GDHC01020914 JAP97714.1 GFPF01007376 MAA18522.1 GEDV01004782 JAP83775.1 CH933809 EDW19311.1 CM002912 KMY99011.1 CM000363 EDX10095.1 CH480815 EDW41120.1 GBXI01017423 JAC96868.1 OUUW01000002 SPP77086.1 X95956 AB176550 AE014296 AY071270 AAF50046.2 AAL48892.1 BAD54754.1 CAA65194.1 ABLF02031480 GFAA01003514 JAT99920.1 KK852432 KDR24004.1 CH479199 EDW29400.1 CM000159 EDW94152.1 CH940647 EDW70930.1 KRF85195.1 GANP01007871 JAB76597.1 GFAC01005082 JAT94106.1 ABJB010465798 ABJB010495453 ABJB010952312 ABJB010978496 DS823764 EEC11850.1 KQ414621 KOC67579.1 CH379069 KK854178 PTY12654.1 CCAG010020780 CH954178 EDV51430.1 CP012525 ALC44725.1 GANP01012886 JAB71582.1 CH902618 EDV39360.1 JH430339 GGLE01002047 MBY06173.1 CH963847 EDW72750.2 GAMC01009462 GAMC01009460 GAMC01009458 GAMC01009456 GAMC01009454 GAMC01009452 JAB97099.1 GDHF01028653 JAI23661.1 CVRI01000037 CRK93645.1 CH916366 EDV97088.1 GEDC01001062 JAS36236.1 UFQT01000995 SSX28355.1 UFQS01001598 UFQT01001598 SSX11541.1 SSX31108.1 KQ761797 OAD56791.1 GAKP01018885 GAKP01018884 JAC40067.1 KQ976440 KYM86491.1 APCN01003494 GDRN01100275 GDRN01100274 GDRN01100273 GDRN01100271 JAI58605.1 AXCM01003724 GDIP01251960 JAI71441.1 AAAB01008979 EAA13728.4 DS231923 EDS26784.1 GDIP01251916 JAI71485.1 AXCN02001805 CAEY01001374 GDIQ01152320 JAK99405.1 GBYB01008837 JAG78604.1
KZ288253 PBC30948.1 LJIG01002331 KRT84609.1 NEVH01019395 PNF22479.1 GL888175 EGI66055.1 KQ982330 KYQ57674.1 KQ971342 EFA03006.2 ADTU01005005 ADTU01005006 GBBI01002782 JAC15930.1 KK107144 QOIP01000003 EZA57599.1 RLU24315.1 KQ977580 KYN01760.1 GL446154 EFN88395.1 KQ981856 KYN34645.1 KQ434777 KZC04285.1 KQ980404 KYN16198.1 BT126988 AEE61950.1 APGK01034125 KB740904 ENN78451.1 NNAY01003486 OXU19345.1 KB632006 ERL87963.1 GEZM01059416 JAV71489.1 DS235219 EEB13374.1 GBYB01008836 GBYB01008838 JAG78603.1 JAG78605.1 GEFH01002408 JAP66173.1 GEDC01016863 GEDC01009346 JAS20435.1 JAS27952.1 GEBQ01028051 GEBQ01003489 JAT11926.1 JAT36488.1 JRES01000005 KNC34982.1 GACK01005177 JAA59857.1 GDHC01020914 JAP97714.1 GFPF01007376 MAA18522.1 GEDV01004782 JAP83775.1 CH933809 EDW19311.1 CM002912 KMY99011.1 CM000363 EDX10095.1 CH480815 EDW41120.1 GBXI01017423 JAC96868.1 OUUW01000002 SPP77086.1 X95956 AB176550 AE014296 AY071270 AAF50046.2 AAL48892.1 BAD54754.1 CAA65194.1 ABLF02031480 GFAA01003514 JAT99920.1 KK852432 KDR24004.1 CH479199 EDW29400.1 CM000159 EDW94152.1 CH940647 EDW70930.1 KRF85195.1 GANP01007871 JAB76597.1 GFAC01005082 JAT94106.1 ABJB010465798 ABJB010495453 ABJB010952312 ABJB010978496 DS823764 EEC11850.1 KQ414621 KOC67579.1 CH379069 KK854178 PTY12654.1 CCAG010020780 CH954178 EDV51430.1 CP012525 ALC44725.1 GANP01012886 JAB71582.1 CH902618 EDV39360.1 JH430339 GGLE01002047 MBY06173.1 CH963847 EDW72750.2 GAMC01009462 GAMC01009460 GAMC01009458 GAMC01009456 GAMC01009454 GAMC01009452 JAB97099.1 GDHF01028653 JAI23661.1 CVRI01000037 CRK93645.1 CH916366 EDV97088.1 GEDC01001062 JAS36236.1 UFQT01000995 SSX28355.1 UFQS01001598 UFQT01001598 SSX11541.1 SSX31108.1 KQ761797 OAD56791.1 GAKP01018885 GAKP01018884 JAC40067.1 KQ976440 KYM86491.1 APCN01003494 GDRN01100275 GDRN01100274 GDRN01100273 GDRN01100271 JAI58605.1 AXCM01003724 GDIP01251960 JAI71441.1 AAAB01008979 EAA13728.4 DS231923 EDS26784.1 GDIP01251916 JAI71485.1 AXCN02001805 CAEY01001374 GDIQ01152320 JAK99405.1 GBYB01008837 JAG78604.1
Proteomes
UP000007151
UP000218220
UP000242457
UP000005203
UP000235965
UP000007755
+ More
UP000075809 UP000007266 UP000005205 UP000053097 UP000279307 UP000078542 UP000008237 UP000078541 UP000002358 UP000076502 UP000078492 UP000019118 UP000215335 UP000030742 UP000009046 UP000037069 UP000009192 UP000000304 UP000001292 UP000268350 UP000000803 UP000007819 UP000027135 UP000008744 UP000002282 UP000078200 UP000091820 UP000008792 UP000001555 UP000053825 UP000001819 UP000092444 UP000008711 UP000092553 UP000007801 UP000007798 UP000183832 UP000095300 UP000001070 UP000078540 UP000075881 UP000075840 UP000192221 UP000075900 UP000075903 UP000076408 UP000075883 UP000076407 UP000007062 UP000075902 UP000075920 UP000075901 UP000002320 UP000075884 UP000075885 UP000075886 UP000015104
UP000075809 UP000007266 UP000005205 UP000053097 UP000279307 UP000078542 UP000008237 UP000078541 UP000002358 UP000076502 UP000078492 UP000019118 UP000215335 UP000030742 UP000009046 UP000037069 UP000009192 UP000000304 UP000001292 UP000268350 UP000000803 UP000007819 UP000027135 UP000008744 UP000002282 UP000078200 UP000091820 UP000008792 UP000001555 UP000053825 UP000001819 UP000092444 UP000008711 UP000092553 UP000007801 UP000007798 UP000183832 UP000095300 UP000001070 UP000078540 UP000075881 UP000075840 UP000192221 UP000075900 UP000075903 UP000076408 UP000075883 UP000076407 UP000007062 UP000075902 UP000075920 UP000075901 UP000002320 UP000075884 UP000075885 UP000075886 UP000015104
Interpro
CDD
ProteinModelPortal
A0A2H1WVF1
A0A212EJ94
A0A2A4J5U6
A0A2A3EGW3
A0A088ATG7
A0A0T6BBD0
+ More
A0A2J7Q1P0 F4WIC4 A0A151XBF5 D6WKM8 A0A158NZU9 A0A023F3C7 A0A026WRA1 A0A195CM25 E2B7B0 A0A195F241 K7INM0 A0A154NX72 A0A151J2J5 J3JUS3 N6UCU0 A0A232ELW1 U4U6M1 A0A1Y1LCU5 E0VJ18 A0A0C9Q470 A0A131XGA6 A0A1B6DQK1 A0A1B6KKG3 A0A0L0CS09 L7M7K2 A0A146KPU2 A0A224YXG8 A0A131YZ60 B4KUQ5 A0A0J9RTB0 B4QQ77 B4HEG6 A0A0A1WD25 A0A3B0JUQ1 Q9VTK2 J9JYV6 A0A1E1XKQ7 A0A067RVV3 B4GZH3 B4PF70 A0A1A9UPF5 A0A1A9WGH5 B4LCN8 V5HMB9 A0A1E1X503 B7PZ28 A0A0L7R9R8 Q2LZ62 A0A2R7VYT2 A0A1B0G158 B3NGX3 A0A0M4F0N3 V5HA07 B3M490 T1JMV6 A0A2R5L9K6 B4MKP4 W8C4P4 A0A0K8UAP7 A0A1J1I054 A0A1I8NS44 A0A1I8NS45 B4J2K3 A0A1B6EE73 A0A336MDJ3 A0A336L2C7 A0A310SJW1 A0A034VD69 A0A195BMH9 A0A182K5A9 A0A182I5S5 A0A1W4W798 A0A182REJ6 A0A182V2U2 A0A182YIW7 A0A0P4VZT5 A0A182MN00 A0A182X0I8 A0A0P4WQZ7 Q7Q222 A0A182UJ28 A0A182W9K5 A0A182S7S1 B0WG88 A0A182NV70 A0A182P9V2 A0A0P4WVR7 A0A182QJ90 T1K338 A0A1S4EX29 A0A0P5MSZ4 A0A0C9R7N7
A0A2J7Q1P0 F4WIC4 A0A151XBF5 D6WKM8 A0A158NZU9 A0A023F3C7 A0A026WRA1 A0A195CM25 E2B7B0 A0A195F241 K7INM0 A0A154NX72 A0A151J2J5 J3JUS3 N6UCU0 A0A232ELW1 U4U6M1 A0A1Y1LCU5 E0VJ18 A0A0C9Q470 A0A131XGA6 A0A1B6DQK1 A0A1B6KKG3 A0A0L0CS09 L7M7K2 A0A146KPU2 A0A224YXG8 A0A131YZ60 B4KUQ5 A0A0J9RTB0 B4QQ77 B4HEG6 A0A0A1WD25 A0A3B0JUQ1 Q9VTK2 J9JYV6 A0A1E1XKQ7 A0A067RVV3 B4GZH3 B4PF70 A0A1A9UPF5 A0A1A9WGH5 B4LCN8 V5HMB9 A0A1E1X503 B7PZ28 A0A0L7R9R8 Q2LZ62 A0A2R7VYT2 A0A1B0G158 B3NGX3 A0A0M4F0N3 V5HA07 B3M490 T1JMV6 A0A2R5L9K6 B4MKP4 W8C4P4 A0A0K8UAP7 A0A1J1I054 A0A1I8NS44 A0A1I8NS45 B4J2K3 A0A1B6EE73 A0A336MDJ3 A0A336L2C7 A0A310SJW1 A0A034VD69 A0A195BMH9 A0A182K5A9 A0A182I5S5 A0A1W4W798 A0A182REJ6 A0A182V2U2 A0A182YIW7 A0A0P4VZT5 A0A182MN00 A0A182X0I8 A0A0P4WQZ7 Q7Q222 A0A182UJ28 A0A182W9K5 A0A182S7S1 B0WG88 A0A182NV70 A0A182P9V2 A0A0P4WVR7 A0A182QJ90 T1K338 A0A1S4EX29 A0A0P5MSZ4 A0A0C9R7N7
PDB
3MAL
E-value=2.58569e-07,
Score=134
Ontologies
PATHWAY
GO
PANTHER
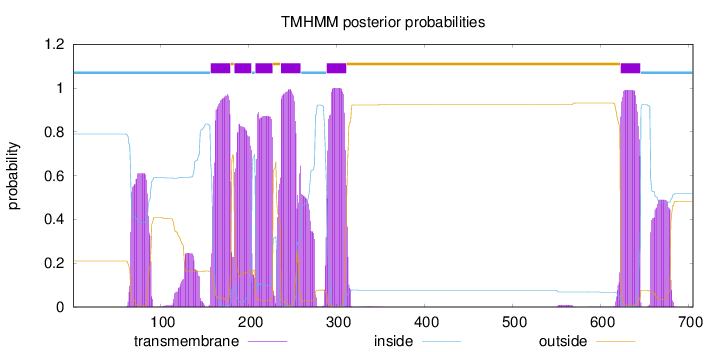
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
705
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
156.67352
Exp number, first 60 AAs:
6e-05
Total prob of N-in:
0.78967
inside
1 - 156
TMhelix
157 - 179
outside
180 - 183
TMhelix
184 - 203
inside
204 - 207
TMhelix
208 - 227
outside
228 - 236
TMhelix
237 - 259
inside
260 - 288
TMhelix
289 - 311
outside
312 - 622
TMhelix
623 - 645
inside
646 - 705
Population Genetic Test Statistics
Pi
178.613657
Theta
178.388492
Tajima's D
0.279474
CLR
1.850559
CSRT
0.454977251137443
Interpretation
Uncertain
