Gene
KWMTBOMO02148 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003009
Annotation
proteasome_subunit_beta_7_[Bombyx_mori]
Full name
Proteasome subunit beta
Location in the cell
Mitochondrial Reliability : 1.768
Sequence
CDS
ATGGCATCTGCATTAGTGCCCGAAATTCCAGTTCCAGGATTCTCGTTTGAGAATTTTCAACGTAATGCTTTTCTTGCTCAGAAAGGGTTTCCTGCACCCAAAGCCACCAAGACCGGTACTACAATTGTTGGTATAATATACGCAGACGGCGTTATTCTAGGAGCTGACACAAGAGCCACTGAAAATACAGTTGTCTCGGACAAAAACTGCCAAAAAATCCACTACTTAGCATCCAACATGTATTGTTGTGGAGCAGGAACAGCAGCTGACACTGAAATGACAACACAATCTGTTGCTTCACAATTGGAGTTGCAGCGCTTACATACTGGACGTACTGTACCTGTAGAAACTGCTGCCACACTGCTGAAACGAATGCTATTTCGCTATCAAGGCCATATAGGGGCTGCTCTGGTTTTGGGAGGTGTTGACAGAACCGGTCCTCATATTTACTGCATTTATCCTCATGGATCTGTAGATAAATTACCATATGCAACCATGGGATCTGGATCTCTTGCGGCTATGGCAGTGTTTGAAGCAGGTTGGAAACGTGATATGAATGAGGAAGAAGGAAAAAAGCTTGTCCGAGATGCTATTGCTGCTGGTATATTCAATGACTTGGGTTCAGGTTCTAACGTTGACCTGTGCGTTATTCGTAACACTGGCCCTGCCCAATATCTAAGAACTTATGAAGAAGCTAATGTGAAGGGGAAAAAGCAAGGTTCGTATAGATATGCTCTTGGTACAACTGCTGTGCTTAAGCAGCGTGTAATACCTCTCGAGGTAGAATCTATTTTGGTACGCCCAACAGAAGCATTTCAGCCTATGGATGTGGAGCCTCAAACGAGTCGTTAA
Protein
MASALVPEIPVPGFSFENFQRNAFLAQKGFPAPKATKTGTTIVGIIYADGVILGADTRATENTVVSDKNCQKIHYLASNMYCCGAGTAADTEMTTQSVASQLELQRLHTGRTVPVETAATLLKRMLFRYQGHIGAALVLGGVDRTGPHIYCIYPHGSVDKLPYATMGSGSLAAMAVFEAGWKRDMNEEEGKKLVRDAIAAGIFNDLGSGSNVDLCVIRNTGPAQYLRTYEEANVKGKKQGSYRYALGTTAVLKQRVIPLEVESILVRPTEAFQPMDVEPQTSR
Summary
Catalytic Activity
Cleavage of peptide bonds with very broad specificity.
Subunit
The 26S proteasome consists of a 20S proteasome core and two 19S regulatory subunits.
Similarity
Belongs to the peptidase T1B family.
Uniprot
Q1HPQ9
I4DJA3
A0A0N0PCC4
A0A2H1WXA3
A0A2A4J5V6
A0A212EJA0
+ More
S4PA70 B6ZAY5 A0A0L7L453 A0A2P8XY49 A0A2J7Q1K5 A0A1Y1LKD8 A0A0T6B1J2 J3JVW5 A0A1L8DT14 A0A1B0DAH0 A0A1W4XGI7 K7IQU2 A0A2A3EIG5 B0WLQ6 A0A232FBJ0 A0A182JY62 A0A088A1E9 D6WKM7 A0A182NPM9 A0A084VII4 A0A182P9F7 A0A1Q3FDV3 A0A182QF53 A0A0K8R709 A0A182RFD2 A0A182IP48 T1E7A1 E0VJR0 A0A182WF28 A0A182YDB0 B7PM02 A0A0K8TS09 K1PVF6 W5J4V1 A0A182VJR1 A0A182UJ83 A0A182XCD1 Q7Q306 A0A182HNN6 A0A2M4AIS8 A0A2M4BW65 Q17FD3 R4UMK4 A0A2M3Z887 A0A182FUV3 A0A182LPL9 A0A2M4AGN9 A0A131XZX4 A0A310SB68 A0A2M3Z8H8 U5ETU2 T1PAN6 A0A182MKH5 A0A026X3R3 A0A023ENF3 E2J7E6 A0A0V0G571 A0A0L7QVX3 T1ID03 A0A0J7KMU4 A0A1B6H327 A0A1I8MT59 E2AHM4 A0A0P4VT79 A0A1L8EEE1 A0A182GK05 A0A1I8P2C4 A0A0M9A4Q0 R4WR55 A0A1L8EED0 A0A0L0C117 E2C832 A0A2R5LHK8 F4WU60 R4FQ45 A0A224XV29 A0A069DRV2 A0A1B6MTX2 A0A158NLU5 A0A0P6JYP6 A0A1E1WV13 A0A1L8EAG0 A0A0K8WL29 A0A034WHR7 A0A1S3JQQ9 A0A154P4M0 A0A0P5H8K7 W8BKV4 A0A0N8DWI8 E9J3A2 E9GG96 C0IXW7 B4KXM3 V4AUS9
S4PA70 B6ZAY5 A0A0L7L453 A0A2P8XY49 A0A2J7Q1K5 A0A1Y1LKD8 A0A0T6B1J2 J3JVW5 A0A1L8DT14 A0A1B0DAH0 A0A1W4XGI7 K7IQU2 A0A2A3EIG5 B0WLQ6 A0A232FBJ0 A0A182JY62 A0A088A1E9 D6WKM7 A0A182NPM9 A0A084VII4 A0A182P9F7 A0A1Q3FDV3 A0A182QF53 A0A0K8R709 A0A182RFD2 A0A182IP48 T1E7A1 E0VJR0 A0A182WF28 A0A182YDB0 B7PM02 A0A0K8TS09 K1PVF6 W5J4V1 A0A182VJR1 A0A182UJ83 A0A182XCD1 Q7Q306 A0A182HNN6 A0A2M4AIS8 A0A2M4BW65 Q17FD3 R4UMK4 A0A2M3Z887 A0A182FUV3 A0A182LPL9 A0A2M4AGN9 A0A131XZX4 A0A310SB68 A0A2M3Z8H8 U5ETU2 T1PAN6 A0A182MKH5 A0A026X3R3 A0A023ENF3 E2J7E6 A0A0V0G571 A0A0L7QVX3 T1ID03 A0A0J7KMU4 A0A1B6H327 A0A1I8MT59 E2AHM4 A0A0P4VT79 A0A1L8EEE1 A0A182GK05 A0A1I8P2C4 A0A0M9A4Q0 R4WR55 A0A1L8EED0 A0A0L0C117 E2C832 A0A2R5LHK8 F4WU60 R4FQ45 A0A224XV29 A0A069DRV2 A0A1B6MTX2 A0A158NLU5 A0A0P6JYP6 A0A1E1WV13 A0A1L8EAG0 A0A0K8WL29 A0A034WHR7 A0A1S3JQQ9 A0A154P4M0 A0A0P5H8K7 W8BKV4 A0A0N8DWI8 E9J3A2 E9GG96 C0IXW7 B4KXM3 V4AUS9
EC Number
3.4.25.1
Pubmed
19121390
22651552
26354079
22118469
23622113
26227816
+ More
29403074 28004739 22516182 23537049 20075255 28648823 18362917 19820115 24438588 20566863 25244985 26369729 22992520 20920257 23761445 12364791 14747013 17210077 17510324 20966253 24508170 30249741 24945155 25315136 20798317 27129103 26483478 23691247 26108605 21719571 26334808 21347285 25348373 24495485 21282665 21292972 19523065 17994087 23254933
29403074 28004739 22516182 23537049 20075255 28648823 18362917 19820115 24438588 20566863 25244985 26369729 22992520 20920257 23761445 12364791 14747013 17210077 17510324 20966253 24508170 30249741 24945155 25315136 20798317 27129103 26483478 23691247 26108605 21719571 26334808 21347285 25348373 24495485 21282665 21292972 19523065 17994087 23254933
EMBL
BABH01010418
DQ443343
ABF51432.1
AK401371
KQ459598
BAM17993.1
+ More
KPI94697.1 KQ460709 KPJ12711.1 ODYU01011353 SOQ57044.1 NWSH01003201 PCG66792.1 AGBW02014506 OWR41555.1 GAIX01003379 JAA89181.1 FJ378902 ACJ13432.1 JTDY01003048 KOB70247.1 PYGN01001174 PSN36942.1 NEVH01019395 PNF22465.1 GEZM01059416 JAV71487.1 LJIG01016322 KRT80949.1 APGK01054294 BT127383 KB741248 KB631794 AEE62345.1 ENN71964.1 ERL86188.1 GFDF01004629 JAV09455.1 AJVK01029070 KZ288250 PBC31072.1 DS231989 EDS30621.1 NNAY01000461 OXU28214.1 KQ971342 EFA03019.1 ATLV01013359 KE524854 KFB37778.1 GFDL01009308 JAV25737.1 AXCN02002112 GADI01006962 JAA66846.1 GAMD01003227 JAA98363.1 DS235226 EEB13616.1 ABJB010456347 ABJB010492253 ABJB011062822 DS744655 EEC07624.1 GDAI01000469 JAI17134.1 JH823227 EKC28337.1 ADMH02002191 ETN57890.1 AAAB01008966 EAA13087.3 APCN01001949 GGFK01007365 MBW40686.1 GGFJ01008175 MBW57316.1 CH477271 EAT45312.1 KC571846 AGM32345.1 GGFM01003960 MBW24711.1 GGFK01006630 MBW39951.1 GEFM01002982 JAP72814.1 KQ765234 OAD54006.1 GGFM01004054 MBW24805.1 GANO01001757 JAB58114.1 KA645802 AFP60431.1 AXCM01011428 KK107019 QOIP01000007 EZA62723.1 RLU20215.1 GAPW01003147 JAC10451.1 HP429364 ADN29864.1 GECL01003586 JAP02538.1 KQ414722 KOC62701.1 ACPB03003020 LBMM01005215 KMQ91708.1 GECZ01000684 JAS69085.1 GL439566 EFN67058.1 GDKW01000888 JAI55707.1 GFDG01001729 JAV17070.1 JXUM01012841 KQ560363 KXJ82808.1 KQ435748 KOX76416.1 AK417142 BAN20357.1 GFDG01001732 JAV17067.1 JRES01001055 KNC25952.1 GL453538 EFN75885.1 GGLE01004860 MBY08986.1 GL888349 EGI62287.1 GAHY01000599 JAA76911.1 GFTR01004473 JAW11953.1 GBGD01002473 JAC86416.1 GEBQ01000607 JAT39370.1 ADTU01019866 ADTU01019867 GDIQ01003248 JAN91489.1 GDQN01000232 JAT90822.1 GFDG01003062 JAV15737.1 GDHF01000537 JAI51777.1 GAKP01004753 JAC54199.1 KQ434819 KZC06885.1 GDIQ01233559 GDIQ01157680 GDIQ01053931 JAK18166.1 GAMC01004620 JAC01936.1 GDIQ01084964 LRGB01000725 JAN09773.1 KZS16309.1 GL768059 EFZ12710.1 GL732543 EFX81362.1 EU564727 ACD87645.1 CH933809 EDW19730.1 KB201205 ESO98720.1
KPI94697.1 KQ460709 KPJ12711.1 ODYU01011353 SOQ57044.1 NWSH01003201 PCG66792.1 AGBW02014506 OWR41555.1 GAIX01003379 JAA89181.1 FJ378902 ACJ13432.1 JTDY01003048 KOB70247.1 PYGN01001174 PSN36942.1 NEVH01019395 PNF22465.1 GEZM01059416 JAV71487.1 LJIG01016322 KRT80949.1 APGK01054294 BT127383 KB741248 KB631794 AEE62345.1 ENN71964.1 ERL86188.1 GFDF01004629 JAV09455.1 AJVK01029070 KZ288250 PBC31072.1 DS231989 EDS30621.1 NNAY01000461 OXU28214.1 KQ971342 EFA03019.1 ATLV01013359 KE524854 KFB37778.1 GFDL01009308 JAV25737.1 AXCN02002112 GADI01006962 JAA66846.1 GAMD01003227 JAA98363.1 DS235226 EEB13616.1 ABJB010456347 ABJB010492253 ABJB011062822 DS744655 EEC07624.1 GDAI01000469 JAI17134.1 JH823227 EKC28337.1 ADMH02002191 ETN57890.1 AAAB01008966 EAA13087.3 APCN01001949 GGFK01007365 MBW40686.1 GGFJ01008175 MBW57316.1 CH477271 EAT45312.1 KC571846 AGM32345.1 GGFM01003960 MBW24711.1 GGFK01006630 MBW39951.1 GEFM01002982 JAP72814.1 KQ765234 OAD54006.1 GGFM01004054 MBW24805.1 GANO01001757 JAB58114.1 KA645802 AFP60431.1 AXCM01011428 KK107019 QOIP01000007 EZA62723.1 RLU20215.1 GAPW01003147 JAC10451.1 HP429364 ADN29864.1 GECL01003586 JAP02538.1 KQ414722 KOC62701.1 ACPB03003020 LBMM01005215 KMQ91708.1 GECZ01000684 JAS69085.1 GL439566 EFN67058.1 GDKW01000888 JAI55707.1 GFDG01001729 JAV17070.1 JXUM01012841 KQ560363 KXJ82808.1 KQ435748 KOX76416.1 AK417142 BAN20357.1 GFDG01001732 JAV17067.1 JRES01001055 KNC25952.1 GL453538 EFN75885.1 GGLE01004860 MBY08986.1 GL888349 EGI62287.1 GAHY01000599 JAA76911.1 GFTR01004473 JAW11953.1 GBGD01002473 JAC86416.1 GEBQ01000607 JAT39370.1 ADTU01019866 ADTU01019867 GDIQ01003248 JAN91489.1 GDQN01000232 JAT90822.1 GFDG01003062 JAV15737.1 GDHF01000537 JAI51777.1 GAKP01004753 JAC54199.1 KQ434819 KZC06885.1 GDIQ01233559 GDIQ01157680 GDIQ01053931 JAK18166.1 GAMC01004620 JAC01936.1 GDIQ01084964 LRGB01000725 JAN09773.1 KZS16309.1 GL768059 EFZ12710.1 GL732543 EFX81362.1 EU564727 ACD87645.1 CH933809 EDW19730.1 KB201205 ESO98720.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000037510
+ More
UP000245037 UP000235965 UP000019118 UP000030742 UP000092462 UP000192223 UP000002358 UP000242457 UP000002320 UP000215335 UP000075881 UP000005203 UP000007266 UP000075884 UP000030765 UP000075885 UP000075886 UP000075900 UP000075880 UP000009046 UP000075920 UP000076408 UP000001555 UP000005408 UP000000673 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000008820 UP000069272 UP000075882 UP000075883 UP000053097 UP000279307 UP000053825 UP000015103 UP000036403 UP000095301 UP000000311 UP000069940 UP000249989 UP000095300 UP000053105 UP000037069 UP000008237 UP000007755 UP000005205 UP000085678 UP000076502 UP000076858 UP000000305 UP000009192 UP000030746
UP000245037 UP000235965 UP000019118 UP000030742 UP000092462 UP000192223 UP000002358 UP000242457 UP000002320 UP000215335 UP000075881 UP000005203 UP000007266 UP000075884 UP000030765 UP000075885 UP000075886 UP000075900 UP000075880 UP000009046 UP000075920 UP000076408 UP000001555 UP000005408 UP000000673 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000008820 UP000069272 UP000075882 UP000075883 UP000053097 UP000279307 UP000053825 UP000015103 UP000036403 UP000095301 UP000000311 UP000069940 UP000249989 UP000095300 UP000053105 UP000037069 UP000008237 UP000007755 UP000005205 UP000085678 UP000076502 UP000076858 UP000000305 UP000009192 UP000030746
Interpro
SUPFAM
SSF56235
SSF56235
Gene 3D
ProteinModelPortal
Q1HPQ9
I4DJA3
A0A0N0PCC4
A0A2H1WXA3
A0A2A4J5V6
A0A212EJA0
+ More
S4PA70 B6ZAY5 A0A0L7L453 A0A2P8XY49 A0A2J7Q1K5 A0A1Y1LKD8 A0A0T6B1J2 J3JVW5 A0A1L8DT14 A0A1B0DAH0 A0A1W4XGI7 K7IQU2 A0A2A3EIG5 B0WLQ6 A0A232FBJ0 A0A182JY62 A0A088A1E9 D6WKM7 A0A182NPM9 A0A084VII4 A0A182P9F7 A0A1Q3FDV3 A0A182QF53 A0A0K8R709 A0A182RFD2 A0A182IP48 T1E7A1 E0VJR0 A0A182WF28 A0A182YDB0 B7PM02 A0A0K8TS09 K1PVF6 W5J4V1 A0A182VJR1 A0A182UJ83 A0A182XCD1 Q7Q306 A0A182HNN6 A0A2M4AIS8 A0A2M4BW65 Q17FD3 R4UMK4 A0A2M3Z887 A0A182FUV3 A0A182LPL9 A0A2M4AGN9 A0A131XZX4 A0A310SB68 A0A2M3Z8H8 U5ETU2 T1PAN6 A0A182MKH5 A0A026X3R3 A0A023ENF3 E2J7E6 A0A0V0G571 A0A0L7QVX3 T1ID03 A0A0J7KMU4 A0A1B6H327 A0A1I8MT59 E2AHM4 A0A0P4VT79 A0A1L8EEE1 A0A182GK05 A0A1I8P2C4 A0A0M9A4Q0 R4WR55 A0A1L8EED0 A0A0L0C117 E2C832 A0A2R5LHK8 F4WU60 R4FQ45 A0A224XV29 A0A069DRV2 A0A1B6MTX2 A0A158NLU5 A0A0P6JYP6 A0A1E1WV13 A0A1L8EAG0 A0A0K8WL29 A0A034WHR7 A0A1S3JQQ9 A0A154P4M0 A0A0P5H8K7 W8BKV4 A0A0N8DWI8 E9J3A2 E9GG96 C0IXW7 B4KXM3 V4AUS9
S4PA70 B6ZAY5 A0A0L7L453 A0A2P8XY49 A0A2J7Q1K5 A0A1Y1LKD8 A0A0T6B1J2 J3JVW5 A0A1L8DT14 A0A1B0DAH0 A0A1W4XGI7 K7IQU2 A0A2A3EIG5 B0WLQ6 A0A232FBJ0 A0A182JY62 A0A088A1E9 D6WKM7 A0A182NPM9 A0A084VII4 A0A182P9F7 A0A1Q3FDV3 A0A182QF53 A0A0K8R709 A0A182RFD2 A0A182IP48 T1E7A1 E0VJR0 A0A182WF28 A0A182YDB0 B7PM02 A0A0K8TS09 K1PVF6 W5J4V1 A0A182VJR1 A0A182UJ83 A0A182XCD1 Q7Q306 A0A182HNN6 A0A2M4AIS8 A0A2M4BW65 Q17FD3 R4UMK4 A0A2M3Z887 A0A182FUV3 A0A182LPL9 A0A2M4AGN9 A0A131XZX4 A0A310SB68 A0A2M3Z8H8 U5ETU2 T1PAN6 A0A182MKH5 A0A026X3R3 A0A023ENF3 E2J7E6 A0A0V0G571 A0A0L7QVX3 T1ID03 A0A0J7KMU4 A0A1B6H327 A0A1I8MT59 E2AHM4 A0A0P4VT79 A0A1L8EEE1 A0A182GK05 A0A1I8P2C4 A0A0M9A4Q0 R4WR55 A0A1L8EED0 A0A0L0C117 E2C832 A0A2R5LHK8 F4WU60 R4FQ45 A0A224XV29 A0A069DRV2 A0A1B6MTX2 A0A158NLU5 A0A0P6JYP6 A0A1E1WV13 A0A1L8EAG0 A0A0K8WL29 A0A034WHR7 A0A1S3JQQ9 A0A154P4M0 A0A0P5H8K7 W8BKV4 A0A0N8DWI8 E9J3A2 E9GG96 C0IXW7 B4KXM3 V4AUS9
PDB
6EPF
E-value=6.2515e-95,
Score=885
Ontologies
GO
PANTHER
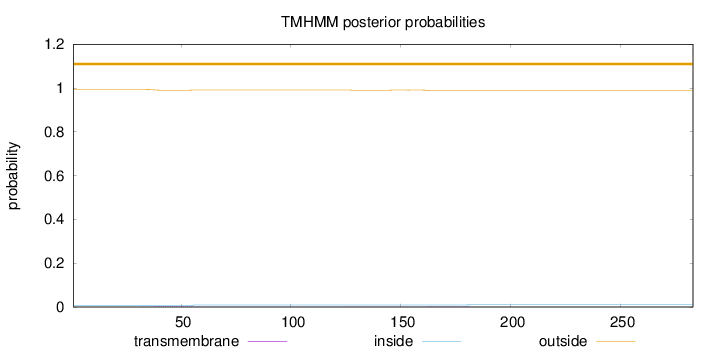
Topology
Subcellular location
Cytoplasm
Nucleus
Nucleus
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11397
Exp number, first 60 AAs:
0.05441
Total prob of N-in:
0.00683
outside
1 - 283
Population Genetic Test Statistics
Pi
139.154197
Theta
125.617202
Tajima's D
0.397168
CLR
0.099347
CSRT
0.482325883705815
Interpretation
Uncertain
