Gene
KWMTBOMO02146
Pre Gene Modal
BGIBMGA003008
Annotation
PREDICTED:_probable_phosphorylase_b_kinase_regulatory_subunit_beta_isoform_X3_[Bombyx_mori]
Full name
Phosphorylase b kinase regulatory subunit
+ More
Probable phosphorylase b kinase regulatory subunit beta
Probable phosphorylase b kinase regulatory subunit beta
Location in the cell
PlasmaMembrane Reliability : 1.974
Sequence
CDS
ATGATTCGTCAAGGAAGCATAGATGACATTAATGCACAACAATATCTAAAGATTTCTAATTATGAAGACACTGTAAGGCAACTTGACATTTGTTATGCAATTGTGAAACGGCAACTTCTACGATTCCAAAGTCCCATAACAGGATTGTTTCCTGTACTATCAACAGAGTTTGAAATAGGAAGTGTGAGGGATAGCATTTACTGTGCTGCAGCTGTTTGGGGTTTATATCAAGCATACAGACGTATTGATGATGACCGAGGTAAATCACATGAACTTGGCCAAAGCACAGTAAAGTGTATGAGAGGCATTTTGGAATGTTGGATCAAACAAGCACCTCGTGTTGAAGCATTCAAGACAAGGCAAAGTGCTGCCCATGCATTACATGTGAAGTTCCACCTTACTACTGGTGAACCGGTCTTGACTGATGACCAATATCATCATTTACAAATTGATGTTGTGTCACTGTATTTACTATTTCTTGTCCAGATGATAACATCTGGCCTACAAATTATATACACTCAGGATGAAGTAGCATTTGTACAGAATCTTGTGTACTATGTTGAACGAGCATATAGAACACCAGACTATGGAATGTGGGAGCGTGGATCTAAATACAATGATGGAAAACCAGAAATCCATGCATCGTCAATAGGTATGGCCAAGGCGGCTCTAGAAGCCATTAATGGTTGTAATTTATTTGGTGATAAGGGTGCATCTTGGAGTGTTGTCTATGTCGACATTGATGCTCACAATCGCAACAGGAGTATATTTGAAACAATGCTACCAAGGGAGTCTAGTTCGAAAGGAGTAGACGCTGCTCTTCTTCCGACCATATCATTTCCTGCTTTTGCCACTCATGAAGAACATTTGGTCCAACTCACCAAAACAAACATAGTAAGACGTTTGCAAGGGAAATATGGTTTTAAGCGTTTTAGTCGTGATGGTTACAAATGTGTTCTCGAAGATCCGGATAGAAGATATTATCATGAAGGGGAATTGAAAAATTTTGATGGTCTAGAATCAGAATGGCCGCTCTTCTATGTTATGATGATTATTGATGGCGTATTTAGAACTTTACCCGACCAAGTTGAAGAGTTTCAGAAGCTTCTGAAAGCTAGGATTTTTATGGACGAACATGGAGATCCTGTAATTCCTTGGTATTACTATATACCCAAAGAAAGTGTTGAAAATGAACGCAATGAACCTAATTCAACTTGGAAAGTACCTTCTACCGGTTTGTTCCTGTGGGGGCAGTCGCTGTTCGTGCTGGCGCAGTTGCTGACCGGGGGCTTGCTGCACGTGAACGAACTCGATCCGATACGAAGGTATCTTCCATCCTACAACCGGCCCCGGCGCGCGGGCCGATATTCCGCTTTTCAGGGCATAGCCACGGATCTGGTTGTTCAAGTGGTACTCATAGCCGAGTCAATGCGTCTACAAGCCATGATGGGTACGTACGGTATACAGACTCAGACACCGCACGAAGTCGAACCCGTTCAGATTTGTAGCTCCACTCAATTAGTTCACGTGTACAGGGAGTTAGGTGTTTGCAAAAAATTGAAGCTGACTGGACGTCCCATTCGACCCATAGGCTCATTGGGTACCAGCAAGATATACAGAGTATGCGGTATGACCGTTTTGTGCTATCCTCTCATTTTCGAAGTTTCCGAGTTTTATCTGTACAGAGACATGGCTTTGCTTATTGATGATATAAAAACTGAACTACAATTCGTTGGGAAATATTGGCGTCTGTCCGGGCGGCCCACCGTGTGCCTGCTGGTTCGGGAAGAGCACATGCGCGACCCACACTTTAAACAGATGTTGGACTTGTTCGCGATGCTGAAGAAGGGCTATTGCGACGGAGTCAAAGTGCGTCTCGGACGATTACAAAATTTAATATCGAGTTCGTGCATGGAACATTTAGACTTTATGAGCCAAGGGGATTTCCCGTCGGAAATGTTTACTCAGTTCAAACAGTTGGAACACGAGTACATAGGCTATCAGTCGCTGACAGACGTTCCACGCACTTTAACCTACCGTGAAGAAACAGTGTCCCTTGAAGCGTACAAACACAAGTCTACTCCTGATGTGGTGGAAGCTCTCAGAACTACTGACAATATATTCGCTCAGTGCCAGTTGTGGGGCATCCTACTCGATAGGGAAGGGCCCATGTATGAAGTGAATGGTACAAACGCTTTGGCGGCGTTGAAGTCCCTGTACCACAGCGCGGGCGGGCTGCGACACTGGCGAGCCGTCCGCTACTGCTCCTCCCTACTGAGCCACACCGTCGATTCCATCAGCCCCTTTATCACTACAGTACTAGTTAACGGGAAACAGTTGACCGTAGGAGTGATAGGACAGAAAGAAACAGTTTTCGACAAACCAATGACTCCGGGAGAGATACAATCCGTAATGTATTCTACCATACAGCCGTATGACATCATAGGCGCAGTATTGCAACAGGAAATAGTGTTGTACTGCGGGCGGCTGATCGGTACGAATCCGGATATATTTCGTGGCATCCTGAAGATCCGCGTGGGCTGGGTGCTGGAGGCCATCCGGTACTACCTGCAGCTGTTCCCCGGCGAGGCGCGGGCCGCGTGCGTCGAGAGCCTGTCCCCCTACCGCCTGCGCACGCTGCTGCAGCGCGTGCTCACCGTGTCCGACTGGGCCGACGAGCGAGGATTAACACCACTACAGCGACGTCAACTGGAAGGCTGCCTCTGTCGAGTTCCTAAACATTTCTACATACAAGTGTGGGATATTTTGCTGCGGACACCTAAAGGAATTATTATTCAGGGCCAGTGTATCCCAGCGGAGCCGACGCTAGTGAACATGTCTCGATCAGAACTGTCGTTTGCTCTGGTAGTGGAAAGCGCCCTCGTTGGTGTGGGAGGGGCGGCGCGGCGGCAGCTGTGCGTGGAGCTGCTGTGCGTGCTCGCCACCATCCTGCGCCGCAACCCCGAGCTGCGCCTGCAGCACTCGCTGGACCTCGACGCGCTGCTGGACGACGCGCACGCCACCTACCGCAAAGACTGCGGGTCCGACGCCGAGCCCGGGCTCAGCACCGCCACGGCCGCGCTCGCCGCCGGACACCTGGCGCGTGCCGTCGTCAACGCCGTGCTGCGCGGCGCCGCCGCTGACCTCGTGCCCGCCGACCGACCCGCCTGTCTCGTCGCCTGA
Protein
MIRQGSIDDINAQQYLKISNYEDTVRQLDICYAIVKRQLLRFQSPITGLFPVLSTEFEIGSVRDSIYCAAAVWGLYQAYRRIDDDRGKSHELGQSTVKCMRGILECWIKQAPRVEAFKTRQSAAHALHVKFHLTTGEPVLTDDQYHHLQIDVVSLYLLFLVQMITSGLQIIYTQDEVAFVQNLVYYVERAYRTPDYGMWERGSKYNDGKPEIHASSIGMAKAALEAINGCNLFGDKGASWSVVYVDIDAHNRNRSIFETMLPRESSSKGVDAALLPTISFPAFATHEEHLVQLTKTNIVRRLQGKYGFKRFSRDGYKCVLEDPDRRYYHEGELKNFDGLESEWPLFYVMMIIDGVFRTLPDQVEEFQKLLKARIFMDEHGDPVIPWYYYIPKESVENERNEPNSTWKVPSTGLFLWGQSLFVLAQLLTGGLLHVNELDPIRRYLPSYNRPRRAGRYSAFQGIATDLVVQVVLIAESMRLQAMMGTYGIQTQTPHEVEPVQICSSTQLVHVYRELGVCKKLKLTGRPIRPIGSLGTSKIYRVCGMTVLCYPLIFEVSEFYLYRDMALLIDDIKTELQFVGKYWRLSGRPTVCLLVREEHMRDPHFKQMLDLFAMLKKGYCDGVKVRLGRLQNLISSSCMEHLDFMSQGDFPSEMFTQFKQLEHEYIGYQSLTDVPRTLTYREETVSLEAYKHKSTPDVVEALRTTDNIFAQCQLWGILLDREGPMYEVNGTNALAALKSLYHSAGGLRHWRAVRYCSSLLSHTVDSISPFITTVLVNGKQLTVGVIGQKETVFDKPMTPGEIQSVMYSTIQPYDIIGAVLQQEIVLYCGRLIGTNPDIFRGILKIRVGWVLEAIRYYLQLFPGEARAACVESLSPYRLRTLLQRVLTVSDWADERGLTPLQRRQLEGCLCRVPKHFYIQVWDILLRTPKGIIIQGQCIPAEPTLVNMSRSELSFALVVESALVGVGGAARRQLCVELLCVLATILRRNPELRLQHSLDLDALLDDAHATYRKDCGSDAEPGLSTATAALAAGHLARAVVNAVLRGAAADLVPADRPACLVA
Summary
Description
Phosphorylase b kinase catalyzes the phosphorylation of serine in certain substrates, including troponin I.
Phosphorylase b kinase catalyzes the phosphorylation of serine in certain substrates, including troponin I. The beta chain acts as a regulatory unit and modulates the activity of the holoenzyme in response to phosphorylation (By similarity).
Phosphorylase b kinase catalyzes the phosphorylation of serine in certain substrates, including troponin I. The beta chain acts as a regulatory unit and modulates the activity of the holoenzyme in response to phosphorylation (By similarity).
Similarity
Belongs to the phosphorylase b kinase regulatory chain family.
Keywords
Alternative splicing
Calmodulin-binding
Carbohydrate metabolism
Cell membrane
Complete proteome
Glycogen metabolism
Lipoprotein
Membrane
Prenylation
Reference proteome
Feature
chain Probable phosphorylase b kinase regulatory subunit beta
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J0H1
A0A194PPN9
A0A0N1I6T3
A0A2A4IU54
A0A1I8QA17
A0A1I8N6K6
+ More
A0A0L0C8W5 A0A1Y1MA72 A0A1I8N6K4 A0A1I8Q9X1 A0A1I8N6L1 A0A1A9YFM7 A0A1I8QA06 A0A2J7RPM1 B4MTZ1 A0A2J7RPM2 A0A1Y1M5A6 A0A1I8N6K5 A0A1A9VXU6 A0A2J7RPM7 Q29KZ6 A0A1I8Q9W9 A0A067R789 A0A1W4V7Q2 B4GSK0 A0A336ME53 A0A0Q9WZZ3 A0A0J9QYE4 A0A3B0JM14 D6WJS2 B3N714 B4NWE7 A0A1B0AGU9 A0A182IQA5 Q9VLS1 B4KGV5 A0A2M3Z187 A0A0R3NU96 A0A182YDP4 B3MLI8 A0A2M4BBD5 A0A2M4A3P1 A0A182RAY4 A0A0R3NU70 A0A1S4H1F7 A0A182UWZ3 A0A182L6F9 K7J3F2 Q7PVH2 A0A2M4CSW1 A0A0Q5WKD8 A0A0A1WEP9 A0A182UHT0 A0A1W4V785 B4LUD0 A0A1A9WMD7 A0A336M4W7 A0A1W4UUX2 B4Q6B0 A0A0J9QZ57 A0A182M501 A0A0P8XFQ7 A0A0C9RGE9 A0A0R1DKJ2 A0A0Q9XCD1 M9PCI5 A0A1W4V8K8 A0A0Q9XGB2 A0A1W4X298 A0A0P8XW49 U5EYS8 B0WBZ1 A0A084VWH0 A0A0Q9X424 B4HYF8 A0A0A1XK07 A0A0Q9XD12 A0A0K8URZ0 A0A0Q9W9S2 W8C3T3 A0A0P8ZFQ2 A0A0Q9X3E1 A0A0C9RTQ7 E2AVZ9 B4JB19 A0A195EFX6 A0A1Q3FKG9 A0A0Q9W9H1 A0A0Q9W9M2 A0A1W4WSM9 A0A195BQ46 A0A026X2T7 A0A151X3A6 A0A232ES65 A0A182HGG0 A0A1W4X3E9 Q176D0 A0A1Q3FKH4 A0A158NTK9 A0A1Q3FKK6
A0A0L0C8W5 A0A1Y1MA72 A0A1I8N6K4 A0A1I8Q9X1 A0A1I8N6L1 A0A1A9YFM7 A0A1I8QA06 A0A2J7RPM1 B4MTZ1 A0A2J7RPM2 A0A1Y1M5A6 A0A1I8N6K5 A0A1A9VXU6 A0A2J7RPM7 Q29KZ6 A0A1I8Q9W9 A0A067R789 A0A1W4V7Q2 B4GSK0 A0A336ME53 A0A0Q9WZZ3 A0A0J9QYE4 A0A3B0JM14 D6WJS2 B3N714 B4NWE7 A0A1B0AGU9 A0A182IQA5 Q9VLS1 B4KGV5 A0A2M3Z187 A0A0R3NU96 A0A182YDP4 B3MLI8 A0A2M4BBD5 A0A2M4A3P1 A0A182RAY4 A0A0R3NU70 A0A1S4H1F7 A0A182UWZ3 A0A182L6F9 K7J3F2 Q7PVH2 A0A2M4CSW1 A0A0Q5WKD8 A0A0A1WEP9 A0A182UHT0 A0A1W4V785 B4LUD0 A0A1A9WMD7 A0A336M4W7 A0A1W4UUX2 B4Q6B0 A0A0J9QZ57 A0A182M501 A0A0P8XFQ7 A0A0C9RGE9 A0A0R1DKJ2 A0A0Q9XCD1 M9PCI5 A0A1W4V8K8 A0A0Q9XGB2 A0A1W4X298 A0A0P8XW49 U5EYS8 B0WBZ1 A0A084VWH0 A0A0Q9X424 B4HYF8 A0A0A1XK07 A0A0Q9XD12 A0A0K8URZ0 A0A0Q9W9S2 W8C3T3 A0A0P8ZFQ2 A0A0Q9X3E1 A0A0C9RTQ7 E2AVZ9 B4JB19 A0A195EFX6 A0A1Q3FKG9 A0A0Q9W9H1 A0A0Q9W9M2 A0A1W4WSM9 A0A195BQ46 A0A026X2T7 A0A151X3A6 A0A232ES65 A0A182HGG0 A0A1W4X3E9 Q176D0 A0A1Q3FKH4 A0A158NTK9 A0A1Q3FKK6
Pubmed
19121390
26354079
25315136
26108605
28004739
17994087
+ More
15632085 24845553 22936249 18362917 19820115 17550304 10731132 12537572 12537569 25244985 12364791 20966253 20075255 25830018 12537568 12537573 12537574 16110336 17569856 17569867 24438588 24495485 20798317 24508170 28648823 17510324 21347285
15632085 24845553 22936249 18362917 19820115 17550304 10731132 12537572 12537569 25244985 12364791 20966253 20075255 25830018 12537568 12537573 12537574 16110336 17569856 17569867 24438588 24495485 20798317 24508170 28648823 17510324 21347285
EMBL
BABH01010419
KQ459598
KPI94699.1
KQ460709
KPJ12709.1
NWSH01006711
+ More
PCG63295.1 JRES01000753 KNC28695.1 GEZM01041629 JAV80247.1 NEVH01001358 PNF42781.1 CH963852 EDW75580.1 PNF42783.1 GEZM01041628 JAV80248.1 PNF42787.1 CH379061 EAL33028.3 KK852829 KDR15354.1 CH479189 EDW25359.1 UFQT01001049 SSX28624.1 KRF98278.1 CM002910 KMY88968.1 OUUW01000006 SPP81903.1 KQ971343 EFA04489.2 CH954177 EDV59310.1 CM000157 EDW88464.1 AE014134 AY089501 CH933807 EDW11155.1 GGFM01001541 MBW22292.1 KRT04679.1 CH902620 EDV31737.1 GGFJ01001224 MBW50365.1 GGFK01002040 MBW35361.1 KRT04680.1 AAAB01008984 EAA14940.4 GGFL01004264 MBW68442.1 KQS70747.1 GBXI01017404 JAC96887.1 CH940649 EDW64116.1 UFQS01000560 UFQT01000560 SSX04954.1 SSX25316.1 SSX28623.1 CM000361 EDX04186.1 KMY88969.1 AXCM01002460 KPU73601.1 GBYB01006116 JAG75883.1 KRJ97831.1 KRG02548.1 AGB92766.1 KRG02547.1 KPU73600.1 GANO01001836 JAB58035.1 DS231883 EDS43005.1 ATLV01017586 KE525174 KFB42314.1 KRG02549.1 CH480818 EDW52088.1 GBXI01003429 JAD10863.1 KRG02546.1 GDHF01022845 JAI29469.1 KRF81434.1 GAMC01009986 JAB96569.1 KPU73599.1 KRG02544.1 GBYB01010946 JAG80713.1 GL443213 EFN62421.1 CH916368 EDW03911.1 KQ978983 KYN26772.1 GFDL01006987 JAV28058.1 KRF81432.1 KRF81433.1 KQ976424 KYM88647.1 KK107020 EZA62413.1 KQ982562 KYQ54871.1 NNAY01002487 OXU21203.1 APCN01004224 CH477390 EAT41971.1 GFDL01007062 JAV27983.1 ADTU01025998 GFDL01006980 JAV28065.1
PCG63295.1 JRES01000753 KNC28695.1 GEZM01041629 JAV80247.1 NEVH01001358 PNF42781.1 CH963852 EDW75580.1 PNF42783.1 GEZM01041628 JAV80248.1 PNF42787.1 CH379061 EAL33028.3 KK852829 KDR15354.1 CH479189 EDW25359.1 UFQT01001049 SSX28624.1 KRF98278.1 CM002910 KMY88968.1 OUUW01000006 SPP81903.1 KQ971343 EFA04489.2 CH954177 EDV59310.1 CM000157 EDW88464.1 AE014134 AY089501 CH933807 EDW11155.1 GGFM01001541 MBW22292.1 KRT04679.1 CH902620 EDV31737.1 GGFJ01001224 MBW50365.1 GGFK01002040 MBW35361.1 KRT04680.1 AAAB01008984 EAA14940.4 GGFL01004264 MBW68442.1 KQS70747.1 GBXI01017404 JAC96887.1 CH940649 EDW64116.1 UFQS01000560 UFQT01000560 SSX04954.1 SSX25316.1 SSX28623.1 CM000361 EDX04186.1 KMY88969.1 AXCM01002460 KPU73601.1 GBYB01006116 JAG75883.1 KRJ97831.1 KRG02548.1 AGB92766.1 KRG02547.1 KPU73600.1 GANO01001836 JAB58035.1 DS231883 EDS43005.1 ATLV01017586 KE525174 KFB42314.1 KRG02549.1 CH480818 EDW52088.1 GBXI01003429 JAD10863.1 KRG02546.1 GDHF01022845 JAI29469.1 KRF81434.1 GAMC01009986 JAB96569.1 KPU73599.1 KRG02544.1 GBYB01010946 JAG80713.1 GL443213 EFN62421.1 CH916368 EDW03911.1 KQ978983 KYN26772.1 GFDL01006987 JAV28058.1 KRF81432.1 KRF81433.1 KQ976424 KYM88647.1 KK107020 EZA62413.1 KQ982562 KYQ54871.1 NNAY01002487 OXU21203.1 APCN01004224 CH477390 EAT41971.1 GFDL01007062 JAV27983.1 ADTU01025998 GFDL01006980 JAV28065.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000095300
UP000095301
+ More
UP000037069 UP000092443 UP000235965 UP000007798 UP000078200 UP000001819 UP000027135 UP000192221 UP000008744 UP000268350 UP000007266 UP000008711 UP000002282 UP000092445 UP000075880 UP000000803 UP000009192 UP000076408 UP000007801 UP000075900 UP000075903 UP000075882 UP000002358 UP000007062 UP000075902 UP000008792 UP000091820 UP000000304 UP000075883 UP000192223 UP000002320 UP000030765 UP000001292 UP000000311 UP000001070 UP000078492 UP000078540 UP000053097 UP000075809 UP000215335 UP000075840 UP000008820 UP000005205
UP000037069 UP000092443 UP000235965 UP000007798 UP000078200 UP000001819 UP000027135 UP000192221 UP000008744 UP000268350 UP000007266 UP000008711 UP000002282 UP000092445 UP000075880 UP000000803 UP000009192 UP000076408 UP000007801 UP000075900 UP000075903 UP000075882 UP000002358 UP000007062 UP000075902 UP000008792 UP000091820 UP000000304 UP000075883 UP000192223 UP000002320 UP000030765 UP000001292 UP000000311 UP000001070 UP000078492 UP000078540 UP000053097 UP000075809 UP000215335 UP000075840 UP000008820 UP000005205
Pfam
PF00723 Glyco_hydro_15
Interpro
SUPFAM
SSF48208
SSF48208
Gene 3D
ProteinModelPortal
H9J0H1
A0A194PPN9
A0A0N1I6T3
A0A2A4IU54
A0A1I8QA17
A0A1I8N6K6
+ More
A0A0L0C8W5 A0A1Y1MA72 A0A1I8N6K4 A0A1I8Q9X1 A0A1I8N6L1 A0A1A9YFM7 A0A1I8QA06 A0A2J7RPM1 B4MTZ1 A0A2J7RPM2 A0A1Y1M5A6 A0A1I8N6K5 A0A1A9VXU6 A0A2J7RPM7 Q29KZ6 A0A1I8Q9W9 A0A067R789 A0A1W4V7Q2 B4GSK0 A0A336ME53 A0A0Q9WZZ3 A0A0J9QYE4 A0A3B0JM14 D6WJS2 B3N714 B4NWE7 A0A1B0AGU9 A0A182IQA5 Q9VLS1 B4KGV5 A0A2M3Z187 A0A0R3NU96 A0A182YDP4 B3MLI8 A0A2M4BBD5 A0A2M4A3P1 A0A182RAY4 A0A0R3NU70 A0A1S4H1F7 A0A182UWZ3 A0A182L6F9 K7J3F2 Q7PVH2 A0A2M4CSW1 A0A0Q5WKD8 A0A0A1WEP9 A0A182UHT0 A0A1W4V785 B4LUD0 A0A1A9WMD7 A0A336M4W7 A0A1W4UUX2 B4Q6B0 A0A0J9QZ57 A0A182M501 A0A0P8XFQ7 A0A0C9RGE9 A0A0R1DKJ2 A0A0Q9XCD1 M9PCI5 A0A1W4V8K8 A0A0Q9XGB2 A0A1W4X298 A0A0P8XW49 U5EYS8 B0WBZ1 A0A084VWH0 A0A0Q9X424 B4HYF8 A0A0A1XK07 A0A0Q9XD12 A0A0K8URZ0 A0A0Q9W9S2 W8C3T3 A0A0P8ZFQ2 A0A0Q9X3E1 A0A0C9RTQ7 E2AVZ9 B4JB19 A0A195EFX6 A0A1Q3FKG9 A0A0Q9W9H1 A0A0Q9W9M2 A0A1W4WSM9 A0A195BQ46 A0A026X2T7 A0A151X3A6 A0A232ES65 A0A182HGG0 A0A1W4X3E9 Q176D0 A0A1Q3FKH4 A0A158NTK9 A0A1Q3FKK6
A0A0L0C8W5 A0A1Y1MA72 A0A1I8N6K4 A0A1I8Q9X1 A0A1I8N6L1 A0A1A9YFM7 A0A1I8QA06 A0A2J7RPM1 B4MTZ1 A0A2J7RPM2 A0A1Y1M5A6 A0A1I8N6K5 A0A1A9VXU6 A0A2J7RPM7 Q29KZ6 A0A1I8Q9W9 A0A067R789 A0A1W4V7Q2 B4GSK0 A0A336ME53 A0A0Q9WZZ3 A0A0J9QYE4 A0A3B0JM14 D6WJS2 B3N714 B4NWE7 A0A1B0AGU9 A0A182IQA5 Q9VLS1 B4KGV5 A0A2M3Z187 A0A0R3NU96 A0A182YDP4 B3MLI8 A0A2M4BBD5 A0A2M4A3P1 A0A182RAY4 A0A0R3NU70 A0A1S4H1F7 A0A182UWZ3 A0A182L6F9 K7J3F2 Q7PVH2 A0A2M4CSW1 A0A0Q5WKD8 A0A0A1WEP9 A0A182UHT0 A0A1W4V785 B4LUD0 A0A1A9WMD7 A0A336M4W7 A0A1W4UUX2 B4Q6B0 A0A0J9QZ57 A0A182M501 A0A0P8XFQ7 A0A0C9RGE9 A0A0R1DKJ2 A0A0Q9XCD1 M9PCI5 A0A1W4V8K8 A0A0Q9XGB2 A0A1W4X298 A0A0P8XW49 U5EYS8 B0WBZ1 A0A084VWH0 A0A0Q9X424 B4HYF8 A0A0A1XK07 A0A0Q9XD12 A0A0K8URZ0 A0A0Q9W9S2 W8C3T3 A0A0P8ZFQ2 A0A0Q9X3E1 A0A0C9RTQ7 E2AVZ9 B4JB19 A0A195EFX6 A0A1Q3FKG9 A0A0Q9W9H1 A0A0Q9W9M2 A0A1W4WSM9 A0A195BQ46 A0A026X2T7 A0A151X3A6 A0A232ES65 A0A182HGG0 A0A1W4X3E9 Q176D0 A0A1Q3FKH4 A0A158NTK9 A0A1Q3FKK6
Ontologies
GO
PANTHER
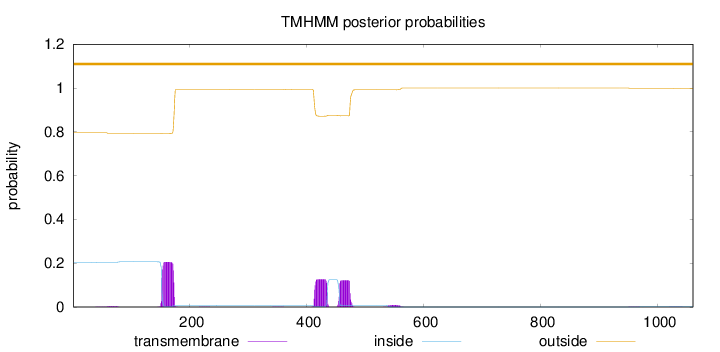
Topology
Subcellular location
Cell membrane
Length:
1060
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
9.81056999999998
Exp number, first 60 AAs:
0.01287
Total prob of N-in:
0.20404
outside
1 - 1060
Population Genetic Test Statistics
Pi
206.99188
Theta
151.276869
Tajima's D
1.39875
CLR
0.30386
CSRT
0.76201189940503
Interpretation
Uncertain
