Gene
KWMTBOMO02141
Pre Gene Modal
BGIBMGA003007
Annotation
PREDICTED:_KDEL_motif-containing_protein_1-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.397 Mitochondrial Reliability : 1.006 PlasmaMembrane Reliability : 1.31
Sequence
CDS
ATGTACAGTTTGTTTATTTTCTTTTTGAAATGGCATTTCATCCAGTGTTCCCAAAACAACGGAGCCGGTATTTATTTGTCAGGACCAGGTCTAAAACCCGATAAGATTGTTTTGCCGGCTAGATATTTCTTCATCACGTTCAGTTCTGTGGATGAAGAGACGTACACTTCAGATTGGGCAAGAAATGTCGCAGTTGAAATTAAAGGAAAAACTCAGAAAACCAACTACTGTAGAATCTGGGTAAATAAATTAGACAGAAAAGATGGGACCTTTATTGTCCGTTACAAAATATATGAAACATGTTTTAATTTGTCTATTAACATTTATTACAAAAGTAAGAATATTGAAGGGTCACCAGTCACATTCAAAGGTCCTATTCTACCAGACCAATGCAACTGTCCAACAAAAGATTTGACGTCATGGTTGACAGACTATGGCTGCAATAATGTACCTGAGCAAATTGTGAATGACCTTAAGGCATTTCAAGCTGTAAATATGACTGTAGAGGTAAAAAGAATTGTAGAGAAGTATCATCAACCGGAAAGCACAAGTTTTTGTCATTACATTATTAAAGGCAATGAAATATTCAGAAATTGCTATGGTCGACATGTGGGTTTTAATATGTTTGCTGACAATATTCTACTATCTCTAACTAGAAAAGTTAAATTGCCTGATGTAGAAATGGTCATAAACTTAGGTGATTGGCCATTAATAAAAAAAAATACAGAACTGTTGCCAGTCTTCTCATGGTGTGGTAGCTCACAGACAATTGATATTCTTATGCCAACCTATGATATTACTGAGTCAACATTAGAGAACATGGGCAGGGTCACTCTGGATATGCTGTCTGTCCAAGGAAATGTGGAACGACCGTGGTCACAACGTGAACCCCGAGCTATTTGGCGAGGTCGAGACTCTCGCTTAGAACGACTTAAACTTGTTGATATTGCTCGATCTCATCCAGATCTGTTTAATGTATCACTTACAAACTTTTTCTTTTTTCGTGAAAAAGAAGAGCAATATGGCCCAAAACAGCCCCACATATCGTTCTTTAAATTCTTTGATTATAAATACCTAATAAATGTGGATGGAACAGTAGCTGCTTATCGTTTCCCATATCTATTAGCTGGAGGAGGACTGGTTTTAAAACAAGACTCTGAATACTATGAACACTTTTACCAGCAACTTCGAGAATGGGAACATTATGTACCAGTTAAAAATGATTTGGGAGACTTAGTAGAAAAAGTTCAGTGGGCTCTCTCGAATGATGACAAAGCACACCAAATTGCAAGAAATGCAAGACAGTTTGCAAATGACCATTTACTTCCACAAAATATAATTTGCTATCATGCTGCTTTGATATCGGAGTGGAGTAAACGAATTTCAAACAAAATTCAAGTCCAAAAAGGTATGACACATGTACCACAGTTAGAATTTGCGTGCGATTGCAGTAACACTGAAGAAGAGATTCACGATGAATTGTGA
Protein
MYSLFIFFLKWHFIQCSQNNGAGIYLSGPGLKPDKIVLPARYFFITFSSVDEETYTSDWARNVAVEIKGKTQKTNYCRIWVNKLDRKDGTFIVRYKIYETCFNLSINIYYKSKNIEGSPVTFKGPILPDQCNCPTKDLTSWLTDYGCNNVPEQIVNDLKAFQAVNMTVEVKRIVEKYHQPESTSFCHYIIKGNEIFRNCYGRHVGFNMFADNILLSLTRKVKLPDVEMVINLGDWPLIKKNTELLPVFSWCGSSQTIDILMPTYDITESTLENMGRVTLDMLSVQGNVERPWSQREPRAIWRGRDSRLERLKLVDIARSHPDLFNVSLTNFFFFREKEEQYGPKQPHISFFKFFDYKYLINVDGTVAAYRFPYLLAGGGLVLKQDSEYYEHFYQQLREWEHYVPVKNDLGDLVEKVQWALSNDDKAHQIARNARQFANDHLLPQNIICYHAALISEWSKRISNKIQVQKGMTHVPQLEFACDCSNTEEEIHDEL
Summary
Uniprot
H9J0H0
A0A0N1IDM4
A0A194PMP3
A0A2H1X1R7
A0A2A4IS92
A0A212FA63
+ More
A0A0L7L9V4 A0A1B6DXF1 A0A067RCK1 A0A1B6FW37 A0A0A9XF34 D6WNM2 A0A023F339 A0A232EXA1 A0A2P2I348 A0A2P8XW28 A0A2J7RL43 A0A088A6E9 A0A1W4W412 A0A0L7R8H9 A0A139WI16 A0A0P6H452 A0A0P6AB01 A0A162P2R9 N6U6C8 A0A1Y1K635 J3JXH0 A0A3L8DRY6 E9G609 A0A147BSQ6 A0A0P4YU30 A0A0P5T591 A0A2A3E360 A0A0P5YNS5 A0A195CNE5 T1HX03 A0A195BKW6 A0A131Z092 A0A158P3W6 F4WKV3 U4TZ54 A0A151J994 A0A151JVH4 U4TUT1 A0A151WVV4 A0A1W4WFU5 A0A293MVP6 A0A0P5VLV6 A0A026VYL1 W8C7F6 A0A2S2QTN5 E2AUD4 A0A1B6DVC0 A0A182MIN3 A0A1W4Z482 A0A182NTR4 A0A182RC75 A0A0C9QD62 A0A182W7B0 A0A182Y016 A0A2M4BKB8 A0A2M4BL59 A0A2M4BK15 A0A2M4BKF1 A0A182P4V2 A0A0A1X683 A0A2H8TZ02 A0A182QZW7 W5JPH1 B4KLU5 A0A084VNE6 A0A034VKF3 A0A154PD06 A0A2S2NLU8 A0A0Q9W4G8 A0A182UST7 Q7Q3T6 A0A182HUX2 A0A0K8W6L3 A0A182WUA4 A0A182F622 A0A1A9Y7N0 A0A3B4AXN4 A0A182U8M8 A0A182JR60 A0A1A9ZW80 A0A1B0C4H6 E0VEQ1 A0A182L1Q3 A0A087TRA6 H2N1M5 A0A1B0GBX2 A0A182HFI4 A0A1A9UNZ2 A0A1W4Z485 A0A1A9X3K4 T1ILJ2 A0A3M6UGD4 B5DSB7 A0A3B3C588
A0A0L7L9V4 A0A1B6DXF1 A0A067RCK1 A0A1B6FW37 A0A0A9XF34 D6WNM2 A0A023F339 A0A232EXA1 A0A2P2I348 A0A2P8XW28 A0A2J7RL43 A0A088A6E9 A0A1W4W412 A0A0L7R8H9 A0A139WI16 A0A0P6H452 A0A0P6AB01 A0A162P2R9 N6U6C8 A0A1Y1K635 J3JXH0 A0A3L8DRY6 E9G609 A0A147BSQ6 A0A0P4YU30 A0A0P5T591 A0A2A3E360 A0A0P5YNS5 A0A195CNE5 T1HX03 A0A195BKW6 A0A131Z092 A0A158P3W6 F4WKV3 U4TZ54 A0A151J994 A0A151JVH4 U4TUT1 A0A151WVV4 A0A1W4WFU5 A0A293MVP6 A0A0P5VLV6 A0A026VYL1 W8C7F6 A0A2S2QTN5 E2AUD4 A0A1B6DVC0 A0A182MIN3 A0A1W4Z482 A0A182NTR4 A0A182RC75 A0A0C9QD62 A0A182W7B0 A0A182Y016 A0A2M4BKB8 A0A2M4BL59 A0A2M4BK15 A0A2M4BKF1 A0A182P4V2 A0A0A1X683 A0A2H8TZ02 A0A182QZW7 W5JPH1 B4KLU5 A0A084VNE6 A0A034VKF3 A0A154PD06 A0A2S2NLU8 A0A0Q9W4G8 A0A182UST7 Q7Q3T6 A0A182HUX2 A0A0K8W6L3 A0A182WUA4 A0A182F622 A0A1A9Y7N0 A0A3B4AXN4 A0A182U8M8 A0A182JR60 A0A1A9ZW80 A0A1B0C4H6 E0VEQ1 A0A182L1Q3 A0A087TRA6 H2N1M5 A0A1B0GBX2 A0A182HFI4 A0A1A9UNZ2 A0A1W4Z485 A0A1A9X3K4 T1ILJ2 A0A3M6UGD4 B5DSB7 A0A3B3C588
Pubmed
19121390
26354079
22118469
26227816
24845553
25401762
+ More
26823975 18362917 19820115 25474469 28648823 29403074 23537049 28004739 22516182 30249741 21292972 29652888 26830274 21347285 21719571 24508170 24495485 20798317 25244985 25830018 20920257 23761445 17994087 24438588 25348373 12364791 14747013 17210077 25463417 20566863 20966253 17554307 26483478 30382153 15632085 29451363
26823975 18362917 19820115 25474469 28648823 29403074 23537049 28004739 22516182 30249741 21292972 29652888 26830274 21347285 21719571 24508170 24495485 20798317 25244985 25830018 20920257 23761445 17994087 24438588 25348373 12364791 14747013 17210077 25463417 20566863 20966253 17554307 26483478 30382153 15632085 29451363
EMBL
BABH01010423
KQ460709
KPJ12707.1
KQ459598
KPI94701.1
ODYU01012783
+ More
SOQ59263.1 NWSH01008529 PCG62519.1 AGBW02009514 OWR50618.1 JTDY01002042 KOB72257.1 GEDC01013091 GEDC01006958 JAS24207.1 JAS30340.1 KK852595 KDR20636.1 GECZ01015354 JAS54415.1 GBHO01026181 GDHC01020254 GDHC01016915 GDHC01000356 JAG17423.1 JAP98374.1 JAQ01714.1 JAQ18273.1 KQ971342 EFA03823.2 GBBI01003171 JAC15541.1 NNAY01001780 OXU22937.1 IACF01002655 LAB68300.1 PYGN01001259 PSN36213.1 NEVH01002699 PNF41553.1 KQ414632 KOC67192.1 KYB27539.1 GDIQ01034966 JAN59771.1 GDIP01031848 JAM71867.1 LRGB01000512 KZS18469.1 APGK01040689 KB740984 ENN76211.1 GEZM01097383 GEZM01097382 JAV54237.1 BT127939 AEE62901.1 QOIP01000005 RLU22619.1 GL732533 EFX85077.1 GEGO01001605 JAR93799.1 GDIP01222692 JAJ00710.1 GDIP01135248 JAL68466.1 KZ288405 PBC26157.1 GDIP01056558 JAM47157.1 KQ977513 KYN02185.1 ACPB03001560 ACPB03001561 KQ976453 KYM85322.1 GEDV01003704 JAP84853.1 ADTU01008607 ADTU01008608 GL888206 EGI65195.1 KB631675 ERL85308.1 KQ979480 KYN21261.1 KQ981701 KYN37476.1 ERL85309.1 KQ982708 KYQ51805.1 GFWV01018662 MAA43390.1 GDIP01112905 JAL90809.1 KK107570 EZA48857.1 GAMC01008041 JAB98514.1 GGMS01011885 MBY81088.1 GL442815 EFN62991.1 GEDC01029237 GEDC01007674 JAS08061.1 JAS29624.1 AXCM01001558 GBYB01012313 JAG82080.1 GGFJ01004260 MBW53401.1 GGFJ01004387 MBW53528.1 GGFJ01004259 MBW53400.1 GGFJ01004386 MBW53527.1 GBXI01008104 JAD06188.1 GFXV01007672 MBW19477.1 AXCN02000801 ADMH02000877 ETN64785.1 CH933808 EDW09755.2 ATLV01014737 KE524984 KFB39490.1 GAKP01015201 JAC43751.1 KQ434874 KZC09703.1 GGMR01005137 MBY17756.1 CH940648 KRF79853.1 AAAB01008964 EAA12302.4 APCN01002480 GDHF01005573 JAI46741.1 JXJN01025481 AAZO01001585 DS235093 EEB11857.1 KK116389 KFM67645.1 CCAG010005900 JXUM01134344 KQ568107 KXJ69136.1 JH430824 RCHS01001612 RMX52564.1 CH475485 EDY70866.2
SOQ59263.1 NWSH01008529 PCG62519.1 AGBW02009514 OWR50618.1 JTDY01002042 KOB72257.1 GEDC01013091 GEDC01006958 JAS24207.1 JAS30340.1 KK852595 KDR20636.1 GECZ01015354 JAS54415.1 GBHO01026181 GDHC01020254 GDHC01016915 GDHC01000356 JAG17423.1 JAP98374.1 JAQ01714.1 JAQ18273.1 KQ971342 EFA03823.2 GBBI01003171 JAC15541.1 NNAY01001780 OXU22937.1 IACF01002655 LAB68300.1 PYGN01001259 PSN36213.1 NEVH01002699 PNF41553.1 KQ414632 KOC67192.1 KYB27539.1 GDIQ01034966 JAN59771.1 GDIP01031848 JAM71867.1 LRGB01000512 KZS18469.1 APGK01040689 KB740984 ENN76211.1 GEZM01097383 GEZM01097382 JAV54237.1 BT127939 AEE62901.1 QOIP01000005 RLU22619.1 GL732533 EFX85077.1 GEGO01001605 JAR93799.1 GDIP01222692 JAJ00710.1 GDIP01135248 JAL68466.1 KZ288405 PBC26157.1 GDIP01056558 JAM47157.1 KQ977513 KYN02185.1 ACPB03001560 ACPB03001561 KQ976453 KYM85322.1 GEDV01003704 JAP84853.1 ADTU01008607 ADTU01008608 GL888206 EGI65195.1 KB631675 ERL85308.1 KQ979480 KYN21261.1 KQ981701 KYN37476.1 ERL85309.1 KQ982708 KYQ51805.1 GFWV01018662 MAA43390.1 GDIP01112905 JAL90809.1 KK107570 EZA48857.1 GAMC01008041 JAB98514.1 GGMS01011885 MBY81088.1 GL442815 EFN62991.1 GEDC01029237 GEDC01007674 JAS08061.1 JAS29624.1 AXCM01001558 GBYB01012313 JAG82080.1 GGFJ01004260 MBW53401.1 GGFJ01004387 MBW53528.1 GGFJ01004259 MBW53400.1 GGFJ01004386 MBW53527.1 GBXI01008104 JAD06188.1 GFXV01007672 MBW19477.1 AXCN02000801 ADMH02000877 ETN64785.1 CH933808 EDW09755.2 ATLV01014737 KE524984 KFB39490.1 GAKP01015201 JAC43751.1 KQ434874 KZC09703.1 GGMR01005137 MBY17756.1 CH940648 KRF79853.1 AAAB01008964 EAA12302.4 APCN01002480 GDHF01005573 JAI46741.1 JXJN01025481 AAZO01001585 DS235093 EEB11857.1 KK116389 KFM67645.1 CCAG010005900 JXUM01134344 KQ568107 KXJ69136.1 JH430824 RCHS01001612 RMX52564.1 CH475485 EDY70866.2
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000027135 UP000007266 UP000215335 UP000245037 UP000235965 UP000005203 UP000192223 UP000053825 UP000076858 UP000019118 UP000279307 UP000000305 UP000242457 UP000078542 UP000015103 UP000078540 UP000005205 UP000007755 UP000030742 UP000078492 UP000078541 UP000075809 UP000053097 UP000000311 UP000075883 UP000192224 UP000075884 UP000075900 UP000075920 UP000076408 UP000075885 UP000075886 UP000000673 UP000009192 UP000030765 UP000076502 UP000008792 UP000075903 UP000007062 UP000075840 UP000076407 UP000069272 UP000092443 UP000261520 UP000075902 UP000075881 UP000092445 UP000092460 UP000009046 UP000075882 UP000054359 UP000001038 UP000092444 UP000069940 UP000249989 UP000078200 UP000091820 UP000275408 UP000001819 UP000261560
UP000027135 UP000007266 UP000215335 UP000245037 UP000235965 UP000005203 UP000192223 UP000053825 UP000076858 UP000019118 UP000279307 UP000000305 UP000242457 UP000078542 UP000015103 UP000078540 UP000005205 UP000007755 UP000030742 UP000078492 UP000078541 UP000075809 UP000053097 UP000000311 UP000075883 UP000192224 UP000075884 UP000075900 UP000075920 UP000076408 UP000075885 UP000075886 UP000000673 UP000009192 UP000030765 UP000076502 UP000008792 UP000075903 UP000007062 UP000075840 UP000076407 UP000069272 UP000092443 UP000261520 UP000075902 UP000075881 UP000092445 UP000092460 UP000009046 UP000075882 UP000054359 UP000001038 UP000092444 UP000069940 UP000249989 UP000078200 UP000091820 UP000275408 UP000001819 UP000261560
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J0H0
A0A0N1IDM4
A0A194PMP3
A0A2H1X1R7
A0A2A4IS92
A0A212FA63
+ More
A0A0L7L9V4 A0A1B6DXF1 A0A067RCK1 A0A1B6FW37 A0A0A9XF34 D6WNM2 A0A023F339 A0A232EXA1 A0A2P2I348 A0A2P8XW28 A0A2J7RL43 A0A088A6E9 A0A1W4W412 A0A0L7R8H9 A0A139WI16 A0A0P6H452 A0A0P6AB01 A0A162P2R9 N6U6C8 A0A1Y1K635 J3JXH0 A0A3L8DRY6 E9G609 A0A147BSQ6 A0A0P4YU30 A0A0P5T591 A0A2A3E360 A0A0P5YNS5 A0A195CNE5 T1HX03 A0A195BKW6 A0A131Z092 A0A158P3W6 F4WKV3 U4TZ54 A0A151J994 A0A151JVH4 U4TUT1 A0A151WVV4 A0A1W4WFU5 A0A293MVP6 A0A0P5VLV6 A0A026VYL1 W8C7F6 A0A2S2QTN5 E2AUD4 A0A1B6DVC0 A0A182MIN3 A0A1W4Z482 A0A182NTR4 A0A182RC75 A0A0C9QD62 A0A182W7B0 A0A182Y016 A0A2M4BKB8 A0A2M4BL59 A0A2M4BK15 A0A2M4BKF1 A0A182P4V2 A0A0A1X683 A0A2H8TZ02 A0A182QZW7 W5JPH1 B4KLU5 A0A084VNE6 A0A034VKF3 A0A154PD06 A0A2S2NLU8 A0A0Q9W4G8 A0A182UST7 Q7Q3T6 A0A182HUX2 A0A0K8W6L3 A0A182WUA4 A0A182F622 A0A1A9Y7N0 A0A3B4AXN4 A0A182U8M8 A0A182JR60 A0A1A9ZW80 A0A1B0C4H6 E0VEQ1 A0A182L1Q3 A0A087TRA6 H2N1M5 A0A1B0GBX2 A0A182HFI4 A0A1A9UNZ2 A0A1W4Z485 A0A1A9X3K4 T1ILJ2 A0A3M6UGD4 B5DSB7 A0A3B3C588
A0A0L7L9V4 A0A1B6DXF1 A0A067RCK1 A0A1B6FW37 A0A0A9XF34 D6WNM2 A0A023F339 A0A232EXA1 A0A2P2I348 A0A2P8XW28 A0A2J7RL43 A0A088A6E9 A0A1W4W412 A0A0L7R8H9 A0A139WI16 A0A0P6H452 A0A0P6AB01 A0A162P2R9 N6U6C8 A0A1Y1K635 J3JXH0 A0A3L8DRY6 E9G609 A0A147BSQ6 A0A0P4YU30 A0A0P5T591 A0A2A3E360 A0A0P5YNS5 A0A195CNE5 T1HX03 A0A195BKW6 A0A131Z092 A0A158P3W6 F4WKV3 U4TZ54 A0A151J994 A0A151JVH4 U4TUT1 A0A151WVV4 A0A1W4WFU5 A0A293MVP6 A0A0P5VLV6 A0A026VYL1 W8C7F6 A0A2S2QTN5 E2AUD4 A0A1B6DVC0 A0A182MIN3 A0A1W4Z482 A0A182NTR4 A0A182RC75 A0A0C9QD62 A0A182W7B0 A0A182Y016 A0A2M4BKB8 A0A2M4BL59 A0A2M4BK15 A0A2M4BKF1 A0A182P4V2 A0A0A1X683 A0A2H8TZ02 A0A182QZW7 W5JPH1 B4KLU5 A0A084VNE6 A0A034VKF3 A0A154PD06 A0A2S2NLU8 A0A0Q9W4G8 A0A182UST7 Q7Q3T6 A0A182HUX2 A0A0K8W6L3 A0A182WUA4 A0A182F622 A0A1A9Y7N0 A0A3B4AXN4 A0A182U8M8 A0A182JR60 A0A1A9ZW80 A0A1B0C4H6 E0VEQ1 A0A182L1Q3 A0A087TRA6 H2N1M5 A0A1B0GBX2 A0A182HFI4 A0A1A9UNZ2 A0A1W4Z485 A0A1A9X3K4 T1ILJ2 A0A3M6UGD4 B5DSB7 A0A3B3C588
PDB
5UB5
E-value=9.78505e-36,
Score=377
Ontologies
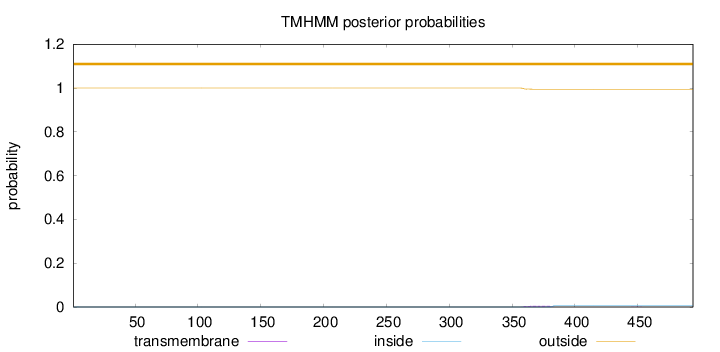
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.517231
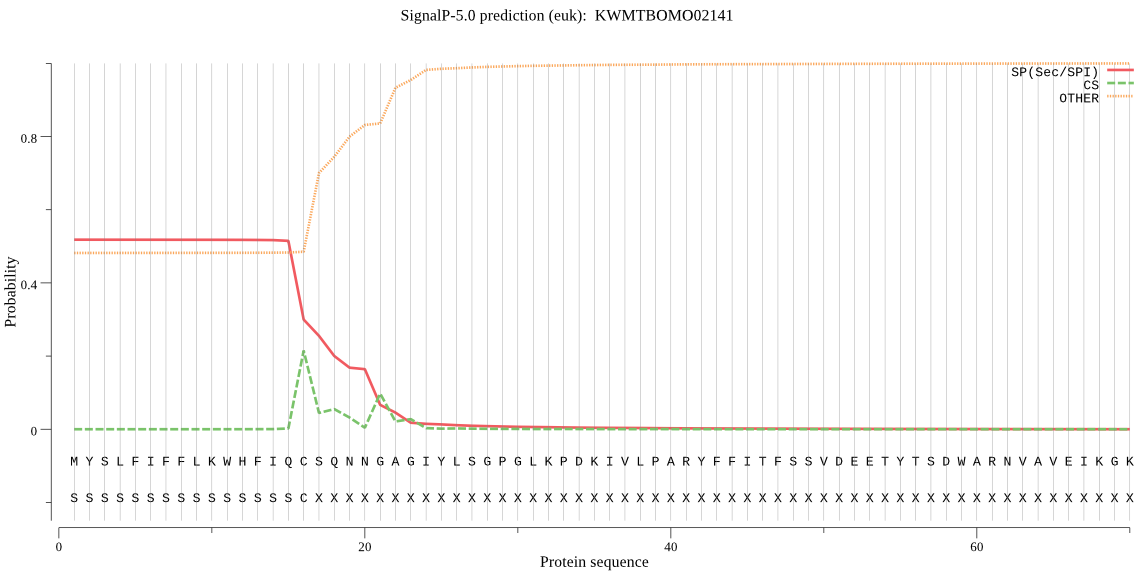
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11617
Exp number, first 60 AAs:
0.00165
Total prob of N-in:
0.00037
outside
1 - 494
Population Genetic Test Statistics
Pi
159.298835
Theta
170.096629
Tajima's D
0.044602
CLR
0.467109
CSRT
0.380280985950702
Interpretation
Uncertain
