Gene
KWMTBOMO02136
Pre Gene Modal
BGIBMGA003005
Annotation
PREDICTED:_repressor_of_RNA_polymerase_III_transcription_MAF1_homolog_[Amyelois_transitella]
Full name
Repressor of RNA polymerase III transcription MAF1
Location in the cell
Extracellular Reliability : 1.121
Sequence
CDS
ATGAAACTATTGGAAAGTGGTCGTTTGGAAGCATTAAGCAGAGCTCTGTCTATTCTGAATGGCGACAGTGCTGTACAAGGCCGCGTTGAAAGCTACAGCTGCAAAATGGCTGGCAGTGAGAAAGCTTTCTATAAGAAGTTTACTGCTGATGGAGAAACTACTCATAGTCTTCAAGCATTGTCTCCCCCTGAGAGTGTTACTTATAGCCGGAGCTTGTCTGGTGACGAAGACGGTGTGTTATGCGACACCATTTCAAGGAAAACATTGTTTTATCTCATTGCTACATTGAATGCTGCTTTTCCAGACTATGATTTCTCCATGGCAAAGAGCAGTGAGTTCAGTGCTGAACCGTCATTGAATTGGGTGCAAAGTGCAGTTGATGCTGCCTTATCTGCAGTAGGTGGTGTGCGTTGGCGCCAACTGCGTCCAGTCTTGTGGGCTGCTGTTGATGAAGAAGTGCTCCTACCAGAATGTCGTATCTACAGCTATAATCCAGACCTAGCTACTGATCCATTTGGTGAACCTGGATGTCTTTGGGCTTACAACTTCTTTTTCTACAATAAGAAGTTGAAAAGGATTGTTTTTCTCACTTTCAGAGCTATAAGCCCTGTTTGTGCAGCCGACTCTGGTGTTGATTTTGCCATGGATGAAGATGAAGATTACAACTGA
Protein
MKLLESGRLEALSRALSILNGDSAVQGRVESYSCKMAGSEKAFYKKFTADGETTHSLQALSPPESVTYSRSLSGDEDGVLCDTISRKTLFYLIATLNAAFPDYDFSMAKSSEFSAEPSLNWVQSAVDAALSAVGGVRWRQLRPVLWAAVDEEVLLPECRIYSYNPDLATDPFGEPGCLWAYNFFFYNKKLKRIVFLTFRAISPVCAADSGVDFAMDEDEDYN
Summary
Description
Element of the TORC1 signaling pathway that acts as a mediator of diverse signals and that represses RNA polymerase III transcription. Inhibits the de novo assembly of TFIIIB onto DNA.
Similarity
Belongs to the MAF1 family.
Uniprot
H9J0G8
A0A1E1W434
A0A0N0PCC1
A0A194PN14
S4PCN0
A0A2A4IWQ6
+ More
A0A2W1BWQ7 A0A2H1WWX4 A0A212FA65 A0A0L7LI19 A0A1Y1N7F8 A0A310S843 A0A154P504 A0A2A3END6 V9IGS1 A0A088A1B7 A0A0N1ITG4 A0A067R0E1 E2BIK6 A0A0L7R5P3 A0A2J7RPH3 A0A232EF08 A0A3L8DA07 V5GQW6 A0A1B6GJW4 A0A1B6JG23 A0A151I7H5 A0A1B6KSD4 R4WE02 A0A0T6B102 A0A2R7X1D4 A0A195EHP3 A0A195FBU5 A0A158P2M4 A0A0A9XZV7 F4W8K0 D6WLB0 A0A151WIU2 A0A2J7RPI4 A0A023FAK2 N6UQ78 T1D4R9 A0A1B0CQ64 A0A195B9Y1 A0A182G688 A0A1S4ENT0 A0A1L8DFR3 A0A0M4EA30 E0W248 W8CCH9 A0A0K8V1W4 A0A034WND9 A0A0A1WY14 B4LQZ7 U5EU43 A0A182WHB5 B4NQN7 A0A0L0CLQ3 A0A1I8PVD4 B4KJ17 R4G5D6 A0A084VMC1 A0A1W4V2U3 T1PF81 A0A0V0G9A7 B3N462 B4IT69 A0A2M4AP38 A0A182M8B2 B4ILX0 A0A0J9S0F3 A0A182NEU6 A0A182QGD7 A0A182F5I9 A0A182IKC8 A0A2M3Z997 W5JD03 Q7PL26 A0A1B0G500 A0A1A9V9H2 A0A1B0A1J4 Q6NL53 A0A1A9XL08 A0A1B0B7P5 B3MWS6 A0A336M6S6 A0A1S4GY73 A0A182IC40 A0A182VDZ5 A0A182XFV5 A0A182L0H0 T1GUE3 E2A3R1 Q174A4 A0A1J1IKH3 A0A023ELZ1 A0A182RG42 A0A1Q3FYC5 A0A0P6IP88 A0A1W4V3A4
A0A2W1BWQ7 A0A2H1WWX4 A0A212FA65 A0A0L7LI19 A0A1Y1N7F8 A0A310S843 A0A154P504 A0A2A3END6 V9IGS1 A0A088A1B7 A0A0N1ITG4 A0A067R0E1 E2BIK6 A0A0L7R5P3 A0A2J7RPH3 A0A232EF08 A0A3L8DA07 V5GQW6 A0A1B6GJW4 A0A1B6JG23 A0A151I7H5 A0A1B6KSD4 R4WE02 A0A0T6B102 A0A2R7X1D4 A0A195EHP3 A0A195FBU5 A0A158P2M4 A0A0A9XZV7 F4W8K0 D6WLB0 A0A151WIU2 A0A2J7RPI4 A0A023FAK2 N6UQ78 T1D4R9 A0A1B0CQ64 A0A195B9Y1 A0A182G688 A0A1S4ENT0 A0A1L8DFR3 A0A0M4EA30 E0W248 W8CCH9 A0A0K8V1W4 A0A034WND9 A0A0A1WY14 B4LQZ7 U5EU43 A0A182WHB5 B4NQN7 A0A0L0CLQ3 A0A1I8PVD4 B4KJ17 R4G5D6 A0A084VMC1 A0A1W4V2U3 T1PF81 A0A0V0G9A7 B3N462 B4IT69 A0A2M4AP38 A0A182M8B2 B4ILX0 A0A0J9S0F3 A0A182NEU6 A0A182QGD7 A0A182F5I9 A0A182IKC8 A0A2M3Z997 W5JD03 Q7PL26 A0A1B0G500 A0A1A9V9H2 A0A1B0A1J4 Q6NL53 A0A1A9XL08 A0A1B0B7P5 B3MWS6 A0A336M6S6 A0A1S4GY73 A0A182IC40 A0A182VDZ5 A0A182XFV5 A0A182L0H0 T1GUE3 E2A3R1 Q174A4 A0A1J1IKH3 A0A023ELZ1 A0A182RG42 A0A1Q3FYC5 A0A0P6IP88 A0A1W4V3A4
Pubmed
19121390
26354079
23622113
28756777
22118469
26227816
+ More
28004739 24845553 20798317 28648823 30249741 23691247 21347285 25401762 26823975 21719571 18362917 19820115 25474469 23537049 24330624 26483478 20566863 24495485 25348373 25830018 17994087 26108605 24438588 25315136 17550304 22936249 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 17510324 24945155
28004739 24845553 20798317 28648823 30249741 23691247 21347285 25401762 26823975 21719571 18362917 19820115 25474469 23537049 24330624 26483478 20566863 24495485 25348373 25830018 17994087 26108605 24438588 25315136 17550304 22936249 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 17510324 24945155
EMBL
BABH01010428
GDQN01009324
JAT81730.1
KQ460709
KPJ12701.1
KQ459598
+ More
KPI94707.1 GAIX01004096 JAA88464.1 NWSH01005404 PCG64245.1 KZ149933 PZC77240.1 ODYU01011624 SOQ57487.1 AGBW02009514 OWR50624.1 JTDY01001039 KOB75050.1 GEZM01010738 JAV93864.1 KQ764881 OAD54321.1 KQ434819 KZC06933.1 KZ288215 PBC32762.1 JR047012 AEY60298.1 KQ435803 KOX73128.1 KK852829 KDR15362.1 GL448516 EFN84453.1 KQ414652 KOC66134.1 NEVH01001358 PNF42730.1 NNAY01005160 OXU16935.1 QOIP01000011 RLU16729.1 GALX01001932 JAB66534.1 GECZ01017450 GECZ01007061 JAS52319.1 JAS62708.1 GECU01036920 GECU01026932 GECU01026021 GECU01025831 GECU01023169 GECU01017863 GECU01015185 GECU01013826 GECU01009475 GECU01008911 GECU01002235 JAS70786.1 JAS80774.1 JAS81685.1 JAS81875.1 JAS84537.1 JAS89843.1 JAS92521.1 JAS93880.1 JAS98231.1 JAS98795.1 JAT05472.1 KQ978406 KYM94149.1 GEBQ01028682 GEBQ01026602 GEBQ01026391 GEBQ01025621 GEBQ01018463 GEBQ01013522 GEBQ01008868 GEBQ01001688 JAT11295.1 JAT13375.1 JAT13586.1 JAT14356.1 JAT21514.1 JAT26455.1 JAT31109.1 JAT38289.1 AK418103 BAN21318.1 LJIG01016299 KRT81018.1 KK856373 PTY25623.1 KQ978881 KYN27775.1 KQ981693 KYN37677.1 ADTU01001276 GBHO01018314 GBHO01018160 GBHO01013187 GBRD01012520 GBRD01008146 GBRD01003005 GDHC01015707 JAG25290.1 JAG25444.1 JAG30417.1 JAG53304.1 JAQ02922.1 GL887908 EGI69491.1 KQ971343 EFA04088.1 KQ983082 KYQ47731.1 PNF42728.1 GBBI01000186 JAC18526.1 APGK01021329 KB740314 KB631743 ENN80892.1 ERL85727.1 GALA01000848 JAA94004.1 AJWK01023163 AJWK01023164 AJWK01023165 KQ976537 KYM81336.1 JXUM01044669 JXUM01044670 JXUM01044671 KQ561409 KXJ78674.1 GFDF01008791 JAV05293.1 CP012523 ALC39176.1 DS235874 EEB19704.1 GAMC01003666 JAC02890.1 GDHF01019392 JAI32922.1 GAKP01003282 JAC55670.1 GBXI01010736 JAD03556.1 CH940649 EDW64536.1 GANO01002486 JAB57385.1 CH964295 EDW86676.1 JRES01000227 KNC33191.1 CH933807 EDW13530.1 ACPB03014855 GAHY01000061 JAA77449.1 ATLV01014585 KE524975 KFB39115.1 KA646552 KA646553 KA647134 KA649721 AFP61182.1 GECL01001589 JAP04535.1 CH954177 EDV59956.1 CH891641 EDW99459.1 GGFK01009224 MBW42545.1 AXCM01000590 CH480903 EDW55925.1 CM002912 KMZ01354.1 AXCN02000840 GGFM01004356 MBW25107.1 ADMH02001866 ETN60775.1 AE013599 EAA46257.2 CCAG010023642 BT012488 AAS93759.1 JXJN01009696 CH902626 EDV41430.1 UFQS01000541 UFQT01000541 SSX04718.1 SSX25081.1 APCN01000774 CAQQ02080973 GL436457 EFN71953.1 CH477415 EAT41366.1 CVRI01000054 CRL00749.1 GAPW01003312 JAC10286.1 GFDL01002461 JAV32584.1 GDIQ01002441 JAN92296.1
KPI94707.1 GAIX01004096 JAA88464.1 NWSH01005404 PCG64245.1 KZ149933 PZC77240.1 ODYU01011624 SOQ57487.1 AGBW02009514 OWR50624.1 JTDY01001039 KOB75050.1 GEZM01010738 JAV93864.1 KQ764881 OAD54321.1 KQ434819 KZC06933.1 KZ288215 PBC32762.1 JR047012 AEY60298.1 KQ435803 KOX73128.1 KK852829 KDR15362.1 GL448516 EFN84453.1 KQ414652 KOC66134.1 NEVH01001358 PNF42730.1 NNAY01005160 OXU16935.1 QOIP01000011 RLU16729.1 GALX01001932 JAB66534.1 GECZ01017450 GECZ01007061 JAS52319.1 JAS62708.1 GECU01036920 GECU01026932 GECU01026021 GECU01025831 GECU01023169 GECU01017863 GECU01015185 GECU01013826 GECU01009475 GECU01008911 GECU01002235 JAS70786.1 JAS80774.1 JAS81685.1 JAS81875.1 JAS84537.1 JAS89843.1 JAS92521.1 JAS93880.1 JAS98231.1 JAS98795.1 JAT05472.1 KQ978406 KYM94149.1 GEBQ01028682 GEBQ01026602 GEBQ01026391 GEBQ01025621 GEBQ01018463 GEBQ01013522 GEBQ01008868 GEBQ01001688 JAT11295.1 JAT13375.1 JAT13586.1 JAT14356.1 JAT21514.1 JAT26455.1 JAT31109.1 JAT38289.1 AK418103 BAN21318.1 LJIG01016299 KRT81018.1 KK856373 PTY25623.1 KQ978881 KYN27775.1 KQ981693 KYN37677.1 ADTU01001276 GBHO01018314 GBHO01018160 GBHO01013187 GBRD01012520 GBRD01008146 GBRD01003005 GDHC01015707 JAG25290.1 JAG25444.1 JAG30417.1 JAG53304.1 JAQ02922.1 GL887908 EGI69491.1 KQ971343 EFA04088.1 KQ983082 KYQ47731.1 PNF42728.1 GBBI01000186 JAC18526.1 APGK01021329 KB740314 KB631743 ENN80892.1 ERL85727.1 GALA01000848 JAA94004.1 AJWK01023163 AJWK01023164 AJWK01023165 KQ976537 KYM81336.1 JXUM01044669 JXUM01044670 JXUM01044671 KQ561409 KXJ78674.1 GFDF01008791 JAV05293.1 CP012523 ALC39176.1 DS235874 EEB19704.1 GAMC01003666 JAC02890.1 GDHF01019392 JAI32922.1 GAKP01003282 JAC55670.1 GBXI01010736 JAD03556.1 CH940649 EDW64536.1 GANO01002486 JAB57385.1 CH964295 EDW86676.1 JRES01000227 KNC33191.1 CH933807 EDW13530.1 ACPB03014855 GAHY01000061 JAA77449.1 ATLV01014585 KE524975 KFB39115.1 KA646552 KA646553 KA647134 KA649721 AFP61182.1 GECL01001589 JAP04535.1 CH954177 EDV59956.1 CH891641 EDW99459.1 GGFK01009224 MBW42545.1 AXCM01000590 CH480903 EDW55925.1 CM002912 KMZ01354.1 AXCN02000840 GGFM01004356 MBW25107.1 ADMH02001866 ETN60775.1 AE013599 EAA46257.2 CCAG010023642 BT012488 AAS93759.1 JXJN01009696 CH902626 EDV41430.1 UFQS01000541 UFQT01000541 SSX04718.1 SSX25081.1 APCN01000774 CAQQ02080973 GL436457 EFN71953.1 CH477415 EAT41366.1 CVRI01000054 CRL00749.1 GAPW01003312 JAC10286.1 GFDL01002461 JAV32584.1 GDIQ01002441 JAN92296.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000037510
+ More
UP000076502 UP000242457 UP000005203 UP000053105 UP000027135 UP000008237 UP000053825 UP000235965 UP000215335 UP000279307 UP000078542 UP000078492 UP000078541 UP000005205 UP000007755 UP000007266 UP000075809 UP000019118 UP000030742 UP000092461 UP000078540 UP000069940 UP000249989 UP000079169 UP000092553 UP000009046 UP000008792 UP000075920 UP000007798 UP000037069 UP000095300 UP000009192 UP000015103 UP000030765 UP000192221 UP000095301 UP000008711 UP000002282 UP000075883 UP000001292 UP000075884 UP000075886 UP000069272 UP000075880 UP000000673 UP000000803 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000007801 UP000075840 UP000075903 UP000076407 UP000075882 UP000015102 UP000000311 UP000008820 UP000183832 UP000075900
UP000076502 UP000242457 UP000005203 UP000053105 UP000027135 UP000008237 UP000053825 UP000235965 UP000215335 UP000279307 UP000078542 UP000078492 UP000078541 UP000005205 UP000007755 UP000007266 UP000075809 UP000019118 UP000030742 UP000092461 UP000078540 UP000069940 UP000249989 UP000079169 UP000092553 UP000009046 UP000008792 UP000075920 UP000007798 UP000037069 UP000095300 UP000009192 UP000015103 UP000030765 UP000192221 UP000095301 UP000008711 UP000002282 UP000075883 UP000001292 UP000075884 UP000075886 UP000069272 UP000075880 UP000000673 UP000000803 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000007801 UP000075840 UP000075903 UP000076407 UP000075882 UP000015102 UP000000311 UP000008820 UP000183832 UP000075900
Gene 3D
ProteinModelPortal
H9J0G8
A0A1E1W434
A0A0N0PCC1
A0A194PN14
S4PCN0
A0A2A4IWQ6
+ More
A0A2W1BWQ7 A0A2H1WWX4 A0A212FA65 A0A0L7LI19 A0A1Y1N7F8 A0A310S843 A0A154P504 A0A2A3END6 V9IGS1 A0A088A1B7 A0A0N1ITG4 A0A067R0E1 E2BIK6 A0A0L7R5P3 A0A2J7RPH3 A0A232EF08 A0A3L8DA07 V5GQW6 A0A1B6GJW4 A0A1B6JG23 A0A151I7H5 A0A1B6KSD4 R4WE02 A0A0T6B102 A0A2R7X1D4 A0A195EHP3 A0A195FBU5 A0A158P2M4 A0A0A9XZV7 F4W8K0 D6WLB0 A0A151WIU2 A0A2J7RPI4 A0A023FAK2 N6UQ78 T1D4R9 A0A1B0CQ64 A0A195B9Y1 A0A182G688 A0A1S4ENT0 A0A1L8DFR3 A0A0M4EA30 E0W248 W8CCH9 A0A0K8V1W4 A0A034WND9 A0A0A1WY14 B4LQZ7 U5EU43 A0A182WHB5 B4NQN7 A0A0L0CLQ3 A0A1I8PVD4 B4KJ17 R4G5D6 A0A084VMC1 A0A1W4V2U3 T1PF81 A0A0V0G9A7 B3N462 B4IT69 A0A2M4AP38 A0A182M8B2 B4ILX0 A0A0J9S0F3 A0A182NEU6 A0A182QGD7 A0A182F5I9 A0A182IKC8 A0A2M3Z997 W5JD03 Q7PL26 A0A1B0G500 A0A1A9V9H2 A0A1B0A1J4 Q6NL53 A0A1A9XL08 A0A1B0B7P5 B3MWS6 A0A336M6S6 A0A1S4GY73 A0A182IC40 A0A182VDZ5 A0A182XFV5 A0A182L0H0 T1GUE3 E2A3R1 Q174A4 A0A1J1IKH3 A0A023ELZ1 A0A182RG42 A0A1Q3FYC5 A0A0P6IP88 A0A1W4V3A4
A0A2W1BWQ7 A0A2H1WWX4 A0A212FA65 A0A0L7LI19 A0A1Y1N7F8 A0A310S843 A0A154P504 A0A2A3END6 V9IGS1 A0A088A1B7 A0A0N1ITG4 A0A067R0E1 E2BIK6 A0A0L7R5P3 A0A2J7RPH3 A0A232EF08 A0A3L8DA07 V5GQW6 A0A1B6GJW4 A0A1B6JG23 A0A151I7H5 A0A1B6KSD4 R4WE02 A0A0T6B102 A0A2R7X1D4 A0A195EHP3 A0A195FBU5 A0A158P2M4 A0A0A9XZV7 F4W8K0 D6WLB0 A0A151WIU2 A0A2J7RPI4 A0A023FAK2 N6UQ78 T1D4R9 A0A1B0CQ64 A0A195B9Y1 A0A182G688 A0A1S4ENT0 A0A1L8DFR3 A0A0M4EA30 E0W248 W8CCH9 A0A0K8V1W4 A0A034WND9 A0A0A1WY14 B4LQZ7 U5EU43 A0A182WHB5 B4NQN7 A0A0L0CLQ3 A0A1I8PVD4 B4KJ17 R4G5D6 A0A084VMC1 A0A1W4V2U3 T1PF81 A0A0V0G9A7 B3N462 B4IT69 A0A2M4AP38 A0A182M8B2 B4ILX0 A0A0J9S0F3 A0A182NEU6 A0A182QGD7 A0A182F5I9 A0A182IKC8 A0A2M3Z997 W5JD03 Q7PL26 A0A1B0G500 A0A1A9V9H2 A0A1B0A1J4 Q6NL53 A0A1A9XL08 A0A1B0B7P5 B3MWS6 A0A336M6S6 A0A1S4GY73 A0A182IC40 A0A182VDZ5 A0A182XFV5 A0A182L0H0 T1GUE3 E2A3R1 Q174A4 A0A1J1IKH3 A0A023ELZ1 A0A182RG42 A0A1Q3FYC5 A0A0P6IP88 A0A1W4V3A4
PDB
3NR5
E-value=8.97635e-43,
Score=434
Ontologies
GO
PANTHER
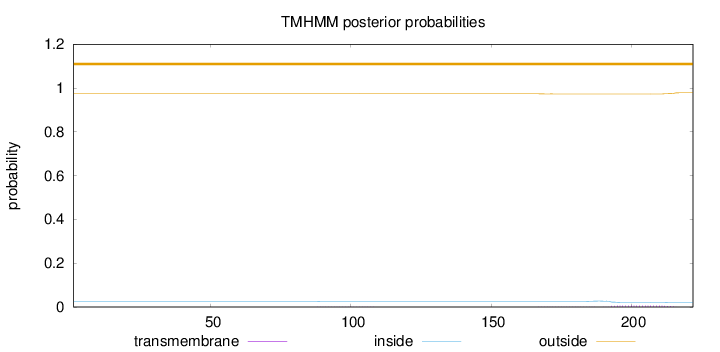
Topology
Subcellular location
Nucleus
Length:
222
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.16743
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02634
outside
1 - 222
Population Genetic Test Statistics
Pi
68.606044
Theta
146.213383
Tajima's D
-0.900938
CLR
442.974302
CSRT
0.155342232888356
Interpretation
Uncertain
