Pre Gene Modal
BGIBMGA003158
Annotation
PREDICTED:_ATP_synthase_mitochondrial_F1_complex_assembly_factor_2_isoform_X1_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.54
Sequence
CDS
ATGAGAATATTTACTCTGGCTTCTGGTATTTCAACTTTACTGAAAACACGATGTACCCTATGCAGTATATATCGTCCATATGCTACACGTAAAAGATTTTATAAGGGGACAGCTGTGATTCAGAATGACAACAAATGGGAAGTGACTTTGGATCATCGACGTCTAAAAACACCAAATGGGAATGTATTAACCGTTGGGAATGAACCTCTTGCTCGTGTTGTTGCTGTAGAATGGGATTCACAAAATGAAACAATTTCACAAGCCACCATGCATTTGACCGCATTGTGTAATACTGCTCTGGACAATCCCGGGAAATTGACATCACATGACATTGTCAATTACCTTTTGGAACATTATCCTACAGACACACTTTTATTTTATTCTGAGGATGAAGAAGAATTGAGGGATCTTCAAGAAAGAAAATGGACACCAATATTGGACTGGTTTCAAAAGACATTTAATGTCAAACAGGAACTAGCGAATAGTATACAGCCACCGCCGGTCTTCACAGAAACTAGAGCTGTGTTAGCTCGGCATTTTCTATCATATGATTTCGCAGCACTGACAGCAATAAATTTCGGTGTCGAGGCATTAAAGTCACCGATCTTAATGTTGGCGTGTATGGAGCGCCGCGTTGTACCCGAGGAAGCTGTATTGTTAGCAAGACTTGAAGAAGAATATCAGCTGGAGCGCTGGGGCCGCGTGCCGTGGGCGCACGGGCTGAGCCAGGCCGACCTGACGGCGCGCGTGTCCGCCGCCCTGCTCCTCGTACACTGCGCTACTGAGAAACATGCCATCAAAACTAAATCTGCCCAAGATTCACAATGA
Protein
MRIFTLASGISTLLKTRCTLCSIYRPYATRKRFYKGTAVIQNDNKWEVTLDHRRLKTPNGNVLTVGNEPLARVVAVEWDSQNETISQATMHLTALCNTALDNPGKLTSHDIVNYLLEHYPTDTLLFYSEDEEELRDLQERKWTPILDWFQKTFNVKQELANSIQPPPVFTETRAVLARHFLSYDFAALTAINFGVEALKSPILMLACMERRVVPEEAVLLARLEEEYQLERWGRVPWAHGLSQADLTARVSAALLLVHCATEKHAIKTKSAQDSQ
Summary
Uniprot
H9J0X1
A0A2A4IY97
A0A2W1BSD4
A0A194PNZ9
S4PI60
A0A212FA51
+ More
D6WNB0 A0A1Y1MF63 A0A1S3D0D6 A0A0N0PC17 A0A067RBP9 N6T7X3 A0A1B6FI91 A0A1B6EXS4 B4KLA4 A0A1B6FBR8 A0A1B6FXK9 A0A1B6DIA5 B4JAY2 A0A2R7WTQ8 A0A1L8E0P2 A0A0M4E9D3 B3MMC5 A0A1L8E0C5 A0A2J7QCP5 A0A336LQI7 E2APH0 B4N7P6 A0A195B0R4 A0A158ND89 A0A151WZX7 A0A0L7R9Y0 A0A1I8PER6 A0A1I8N7E9 F4X763 A0A195DJ90 A0A034VLI9 A0A088AD92 Q29KM5 A0A0K8UC73 A0A3L8DF23 A0A2A3ECK6 B0WBZ8 A0A195FGV9 B4LRW5 W8BUK2 A0A026WHM7 B4GT38 B3NLY0 A0A0A1X1A2 Q9VID7 J3JXB6 A0A1Q3FBY6 B4Q4C9 A0A1B6LWQ7 Q8T3M6 A0A0L0C6S5 A0A154P2F6 A0A310SNW1 A0A1W4UYI2 A0A0Q9WJ30 A0A3B0JV80 E9FRB1 B4IFN0 A0A151I7U0 A0A2M3ZBX9 A0A0A9WGC8 W5J2Z2 A0A2M3ZBT9 B4P6X4 A0A2M4AXQ3 A0A1B0FPC3 Q16TS9 E9J2E3 E0VF77 A0A182H6I9 A0A084VYM6 A0A1A9UH29 A0A1A9XT58 A0A1B0BDE4 A0A182RUR0 A0A1B0DHV8 A0A2M4BWM1 A0A182V534 A0A182XDD4 A0A182LMZ4 Q7PWR4 A0A182I767 A0A0P5NVH8 A0A182U998 A0A232FJ16 A0A0P5VPU1 A0A0P6EQU7 A0A0P5C7Z1 A0A182VU98 A0A0P6C2L6 A0A0P5QFT5 A0A1A9WZH0 T1J1S3
D6WNB0 A0A1Y1MF63 A0A1S3D0D6 A0A0N0PC17 A0A067RBP9 N6T7X3 A0A1B6FI91 A0A1B6EXS4 B4KLA4 A0A1B6FBR8 A0A1B6FXK9 A0A1B6DIA5 B4JAY2 A0A2R7WTQ8 A0A1L8E0P2 A0A0M4E9D3 B3MMC5 A0A1L8E0C5 A0A2J7QCP5 A0A336LQI7 E2APH0 B4N7P6 A0A195B0R4 A0A158ND89 A0A151WZX7 A0A0L7R9Y0 A0A1I8PER6 A0A1I8N7E9 F4X763 A0A195DJ90 A0A034VLI9 A0A088AD92 Q29KM5 A0A0K8UC73 A0A3L8DF23 A0A2A3ECK6 B0WBZ8 A0A195FGV9 B4LRW5 W8BUK2 A0A026WHM7 B4GT38 B3NLY0 A0A0A1X1A2 Q9VID7 J3JXB6 A0A1Q3FBY6 B4Q4C9 A0A1B6LWQ7 Q8T3M6 A0A0L0C6S5 A0A154P2F6 A0A310SNW1 A0A1W4UYI2 A0A0Q9WJ30 A0A3B0JV80 E9FRB1 B4IFN0 A0A151I7U0 A0A2M3ZBX9 A0A0A9WGC8 W5J2Z2 A0A2M3ZBT9 B4P6X4 A0A2M4AXQ3 A0A1B0FPC3 Q16TS9 E9J2E3 E0VF77 A0A182H6I9 A0A084VYM6 A0A1A9UH29 A0A1A9XT58 A0A1B0BDE4 A0A182RUR0 A0A1B0DHV8 A0A2M4BWM1 A0A182V534 A0A182XDD4 A0A182LMZ4 Q7PWR4 A0A182I767 A0A0P5NVH8 A0A182U998 A0A232FJ16 A0A0P5VPU1 A0A0P6EQU7 A0A0P5C7Z1 A0A182VU98 A0A0P6C2L6 A0A0P5QFT5 A0A1A9WZH0 T1J1S3
Pubmed
19121390
28756777
26354079
23622113
22118469
18362917
+ More
19820115 28004739 24845553 23537049 17994087 20798317 21347285 25315136 21719571 25348373 15632085 30249741 24495485 24508170 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 22936249 26108605 21292972 25401762 26823975 20920257 23761445 17550304 17510324 21282665 20566863 26483478 24438588 20966253 12364791 14747013 17210077 28648823
19820115 28004739 24845553 23537049 17994087 20798317 21347285 25315136 21719571 25348373 15632085 30249741 24495485 24508170 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 22936249 26108605 21292972 25401762 26823975 20920257 23761445 17550304 17510324 21282665 20566863 26483478 24438588 20966253 12364791 14747013 17210077 28648823
EMBL
BABH01010433
BABH01010434
NWSH01005404
PCG64244.1
KZ149933
PZC77241.1
+ More
KQ459598 KPI94708.1 GAIX01000279 JAA92281.1 AGBW02009514 OWR50625.1 KQ971342 EFA03810.1 GEZM01032991 JAV84469.1 KQ460709 KPJ12700.1 KK852561 KDR21316.1 APGK01040876 KB740984 KB631899 ENN76329.1 ERL86977.1 GECZ01019850 JAS49919.1 GECZ01027286 JAS42483.1 CH933807 EDW11765.1 GECZ01022171 JAS47598.1 GECZ01014880 JAS54889.1 GEDC01011921 JAS25377.1 CH916368 EDW02852.1 KK855521 PTY22944.1 GFDF01001868 JAV12216.1 CP012523 ALC39727.1 CH902620 EDV31885.2 GFDF01001891 JAV12193.1 NEVH01016288 PNF26343.1 UFQS01000114 UFQT01000114 SSW99840.1 SSX20220.1 GL441542 EFN64669.1 CH964214 EDW80385.1 KQ976690 KYM78051.1 ADTU01012511 KQ982626 KYQ53460.1 KQ414620 KOC67644.1 GL888828 EGI57675.1 KQ980800 KYN12887.1 GAKP01016287 GAKP01016286 JAC42665.1 CH379061 EAL33149.1 GDHF01028072 JAI24242.1 QOIP01000009 RLU18489.1 KZ288285 PBC29467.1 DS231884 EDS43058.1 KQ981560 KYN39920.1 CH940649 EDW63641.2 GAMC01003673 JAC02883.1 KK107199 EZA55562.1 CH479189 EDW25547.1 CH954179 EDV54446.1 GBXI01009742 JAD04550.1 AE014134 AAF53985.1 AGB93203.1 BT127885 AEE62847.1 GFDL01010016 JAV25029.1 CM000361 CM002910 EDX05726.1 KMY91322.1 GEBQ01011893 JAT28084.1 AY094758 AAM11111.1 JRES01000933 KNC27139.1 KQ434787 KZC05300.1 KQ760551 OAD59830.1 KRF81220.1 OUUW01000010 SPP86005.1 GL732523 EFX90134.1 CH480833 EDW46452.1 KQ978392 KYM94269.1 GGFM01005224 MBW25975.1 GBHO01037148 GBHO01037147 GDHC01002488 JAG06456.1 JAG06457.1 JAQ16141.1 ADMH02002160 ETN58186.1 GGFM01005221 MBW25972.1 CM000158 EDW89943.1 GGFK01012243 MBW45564.1 CCAG010019974 CH477641 EAT37913.1 GL767845 EFZ12969.1 DS235107 EEB12033.1 JXUM01114158 KQ565759 KXJ70694.1 ATLV01018379 KE525231 KFB43070.1 JXJN01012447 AJVK01061659 GGFJ01008318 MBW57459.1 AAAB01008984 EAA14894.5 APCN01003368 GDIQ01137137 JAL14589.1 NNAY01000176 OXU30297.1 GDIP01097113 GDIP01097111 JAM06602.1 GDIQ01059622 JAN35115.1 GDIP01174850 GDIP01112209 GDIP01090411 GDIP01045823 GDIP01045821 JAJ48552.1 GDIP01022080 JAM81635.1 GDIQ01117748 JAL33978.1 JH431789
KQ459598 KPI94708.1 GAIX01000279 JAA92281.1 AGBW02009514 OWR50625.1 KQ971342 EFA03810.1 GEZM01032991 JAV84469.1 KQ460709 KPJ12700.1 KK852561 KDR21316.1 APGK01040876 KB740984 KB631899 ENN76329.1 ERL86977.1 GECZ01019850 JAS49919.1 GECZ01027286 JAS42483.1 CH933807 EDW11765.1 GECZ01022171 JAS47598.1 GECZ01014880 JAS54889.1 GEDC01011921 JAS25377.1 CH916368 EDW02852.1 KK855521 PTY22944.1 GFDF01001868 JAV12216.1 CP012523 ALC39727.1 CH902620 EDV31885.2 GFDF01001891 JAV12193.1 NEVH01016288 PNF26343.1 UFQS01000114 UFQT01000114 SSW99840.1 SSX20220.1 GL441542 EFN64669.1 CH964214 EDW80385.1 KQ976690 KYM78051.1 ADTU01012511 KQ982626 KYQ53460.1 KQ414620 KOC67644.1 GL888828 EGI57675.1 KQ980800 KYN12887.1 GAKP01016287 GAKP01016286 JAC42665.1 CH379061 EAL33149.1 GDHF01028072 JAI24242.1 QOIP01000009 RLU18489.1 KZ288285 PBC29467.1 DS231884 EDS43058.1 KQ981560 KYN39920.1 CH940649 EDW63641.2 GAMC01003673 JAC02883.1 KK107199 EZA55562.1 CH479189 EDW25547.1 CH954179 EDV54446.1 GBXI01009742 JAD04550.1 AE014134 AAF53985.1 AGB93203.1 BT127885 AEE62847.1 GFDL01010016 JAV25029.1 CM000361 CM002910 EDX05726.1 KMY91322.1 GEBQ01011893 JAT28084.1 AY094758 AAM11111.1 JRES01000933 KNC27139.1 KQ434787 KZC05300.1 KQ760551 OAD59830.1 KRF81220.1 OUUW01000010 SPP86005.1 GL732523 EFX90134.1 CH480833 EDW46452.1 KQ978392 KYM94269.1 GGFM01005224 MBW25975.1 GBHO01037148 GBHO01037147 GDHC01002488 JAG06456.1 JAG06457.1 JAQ16141.1 ADMH02002160 ETN58186.1 GGFM01005221 MBW25972.1 CM000158 EDW89943.1 GGFK01012243 MBW45564.1 CCAG010019974 CH477641 EAT37913.1 GL767845 EFZ12969.1 DS235107 EEB12033.1 JXUM01114158 KQ565759 KXJ70694.1 ATLV01018379 KE525231 KFB43070.1 JXJN01012447 AJVK01061659 GGFJ01008318 MBW57459.1 AAAB01008984 EAA14894.5 APCN01003368 GDIQ01137137 JAL14589.1 NNAY01000176 OXU30297.1 GDIP01097113 GDIP01097111 JAM06602.1 GDIQ01059622 JAN35115.1 GDIP01174850 GDIP01112209 GDIP01090411 GDIP01045823 GDIP01045821 JAJ48552.1 GDIP01022080 JAM81635.1 GDIQ01117748 JAL33978.1 JH431789
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000007266
UP000079169
+ More
UP000053240 UP000027135 UP000019118 UP000030742 UP000009192 UP000001070 UP000092553 UP000007801 UP000235965 UP000000311 UP000007798 UP000078540 UP000005205 UP000075809 UP000053825 UP000095300 UP000095301 UP000007755 UP000078492 UP000005203 UP000001819 UP000279307 UP000242457 UP000002320 UP000078541 UP000008792 UP000053097 UP000008744 UP000008711 UP000000803 UP000000304 UP000037069 UP000076502 UP000192221 UP000268350 UP000000305 UP000001292 UP000078542 UP000000673 UP000002282 UP000092444 UP000008820 UP000009046 UP000069940 UP000249989 UP000030765 UP000078200 UP000092443 UP000092460 UP000075900 UP000092462 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000215335 UP000075920 UP000091820
UP000053240 UP000027135 UP000019118 UP000030742 UP000009192 UP000001070 UP000092553 UP000007801 UP000235965 UP000000311 UP000007798 UP000078540 UP000005205 UP000075809 UP000053825 UP000095300 UP000095301 UP000007755 UP000078492 UP000005203 UP000001819 UP000279307 UP000242457 UP000002320 UP000078541 UP000008792 UP000053097 UP000008744 UP000008711 UP000000803 UP000000304 UP000037069 UP000076502 UP000192221 UP000268350 UP000000305 UP000001292 UP000078542 UP000000673 UP000002282 UP000092444 UP000008820 UP000009046 UP000069940 UP000249989 UP000030765 UP000078200 UP000092443 UP000092460 UP000075900 UP000092462 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000215335 UP000075920 UP000091820
Pfam
PF07542 ATP12
Interpro
Gene 3D
ProteinModelPortal
H9J0X1
A0A2A4IY97
A0A2W1BSD4
A0A194PNZ9
S4PI60
A0A212FA51
+ More
D6WNB0 A0A1Y1MF63 A0A1S3D0D6 A0A0N0PC17 A0A067RBP9 N6T7X3 A0A1B6FI91 A0A1B6EXS4 B4KLA4 A0A1B6FBR8 A0A1B6FXK9 A0A1B6DIA5 B4JAY2 A0A2R7WTQ8 A0A1L8E0P2 A0A0M4E9D3 B3MMC5 A0A1L8E0C5 A0A2J7QCP5 A0A336LQI7 E2APH0 B4N7P6 A0A195B0R4 A0A158ND89 A0A151WZX7 A0A0L7R9Y0 A0A1I8PER6 A0A1I8N7E9 F4X763 A0A195DJ90 A0A034VLI9 A0A088AD92 Q29KM5 A0A0K8UC73 A0A3L8DF23 A0A2A3ECK6 B0WBZ8 A0A195FGV9 B4LRW5 W8BUK2 A0A026WHM7 B4GT38 B3NLY0 A0A0A1X1A2 Q9VID7 J3JXB6 A0A1Q3FBY6 B4Q4C9 A0A1B6LWQ7 Q8T3M6 A0A0L0C6S5 A0A154P2F6 A0A310SNW1 A0A1W4UYI2 A0A0Q9WJ30 A0A3B0JV80 E9FRB1 B4IFN0 A0A151I7U0 A0A2M3ZBX9 A0A0A9WGC8 W5J2Z2 A0A2M3ZBT9 B4P6X4 A0A2M4AXQ3 A0A1B0FPC3 Q16TS9 E9J2E3 E0VF77 A0A182H6I9 A0A084VYM6 A0A1A9UH29 A0A1A9XT58 A0A1B0BDE4 A0A182RUR0 A0A1B0DHV8 A0A2M4BWM1 A0A182V534 A0A182XDD4 A0A182LMZ4 Q7PWR4 A0A182I767 A0A0P5NVH8 A0A182U998 A0A232FJ16 A0A0P5VPU1 A0A0P6EQU7 A0A0P5C7Z1 A0A182VU98 A0A0P6C2L6 A0A0P5QFT5 A0A1A9WZH0 T1J1S3
D6WNB0 A0A1Y1MF63 A0A1S3D0D6 A0A0N0PC17 A0A067RBP9 N6T7X3 A0A1B6FI91 A0A1B6EXS4 B4KLA4 A0A1B6FBR8 A0A1B6FXK9 A0A1B6DIA5 B4JAY2 A0A2R7WTQ8 A0A1L8E0P2 A0A0M4E9D3 B3MMC5 A0A1L8E0C5 A0A2J7QCP5 A0A336LQI7 E2APH0 B4N7P6 A0A195B0R4 A0A158ND89 A0A151WZX7 A0A0L7R9Y0 A0A1I8PER6 A0A1I8N7E9 F4X763 A0A195DJ90 A0A034VLI9 A0A088AD92 Q29KM5 A0A0K8UC73 A0A3L8DF23 A0A2A3ECK6 B0WBZ8 A0A195FGV9 B4LRW5 W8BUK2 A0A026WHM7 B4GT38 B3NLY0 A0A0A1X1A2 Q9VID7 J3JXB6 A0A1Q3FBY6 B4Q4C9 A0A1B6LWQ7 Q8T3M6 A0A0L0C6S5 A0A154P2F6 A0A310SNW1 A0A1W4UYI2 A0A0Q9WJ30 A0A3B0JV80 E9FRB1 B4IFN0 A0A151I7U0 A0A2M3ZBX9 A0A0A9WGC8 W5J2Z2 A0A2M3ZBT9 B4P6X4 A0A2M4AXQ3 A0A1B0FPC3 Q16TS9 E9J2E3 E0VF77 A0A182H6I9 A0A084VYM6 A0A1A9UH29 A0A1A9XT58 A0A1B0BDE4 A0A182RUR0 A0A1B0DHV8 A0A2M4BWM1 A0A182V534 A0A182XDD4 A0A182LMZ4 Q7PWR4 A0A182I767 A0A0P5NVH8 A0A182U998 A0A232FJ16 A0A0P5VPU1 A0A0P6EQU7 A0A0P5C7Z1 A0A182VU98 A0A0P6C2L6 A0A0P5QFT5 A0A1A9WZH0 T1J1S3
PDB
2R31
E-value=2.20811e-13,
Score=181
Ontologies
GO
PANTHER
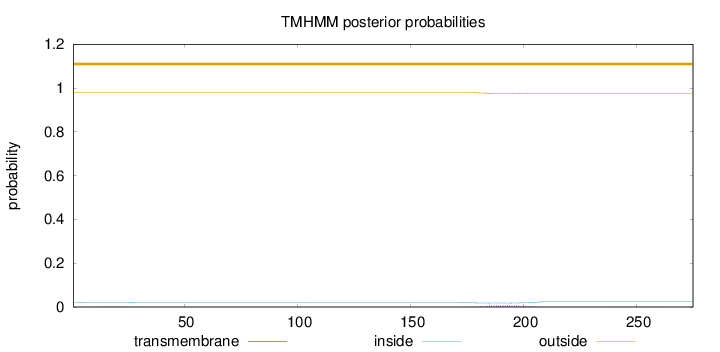
Topology
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2064
Exp number, first 60 AAs:
0.0283
Total prob of N-in:
0.01992
outside
1 - 275
Population Genetic Test Statistics
Pi
246.548773
Theta
173.33498
Tajima's D
1.736473
CLR
0.541566
CSRT
0.833458327083646
Interpretation
Uncertain
