Gene
KWMTBOMO02124
Annotation
PREDICTED:_hydrocephalus-inducing_protein_homolog_[Bombyx_mori]
Full name
Hydrocephalus-inducing protein
+ More
Hydrocephalus-inducing protein homolog
Hydrocephalus-inducing protein homolog
Alternative Name
Protein Hy-3
Location in the cell
Cytoplasmic Reliability : 2.752
Sequence
CDS
ATGTCGTACTCGCCATTAGTGGTCGGCGAGAGGGAGGATACATTGGAAATCTACAATAACTTAGTTGGCACTTACATATACAGAGTGAAGCTTAAGTGTCTCCCGGGCAAAGAGAAGAACCTTGAATTCACGACAAGTTTTGGTACTTGTATCCCTTTACGGCTTCGAATACAGAATAAAACCGATGTGAAAACAGATTTCATAGCTGAGGTATCTCATTCTTCAATTCAAATAGATAAAGAATGTTGCTTGGGACCATTAGAAAAAGGAAAGATTTTAGTGTGGTTTGAACCAACACAGTTGGGAGTGCAGAATTGTAAAGTGTCCTTCAAATCGACGCTCGCTGGTGAATTTGTATTTAATATTAAAGGAATAGCGACGGAACCCAAGCCGCAGGGCCCGTACGAAGTTAAGGCTGGTGGATCTACAACTATTAAATTTAAAAATGTCTTCGATTCATCGAGAATGTTCAAAATATACGTAGACCGGGAGGAGTTTTACGTTAAAACTATGTTTGAAACTATAAAACCAAAGAAGGACCTGAGAATATCTGTTTATCTGAACGACAAGCCCGCACAAGGCTGGACAGAAGTACCTACTGGATGTTTAACTATAGAGACACACGAACCAGCTGAACCTAAAGTACACTGGACTTATTTTCTACAGGGAAAACAGTAG
Protein
MSYSPLVVGEREDTLEIYNNLVGTYIYRVKLKCLPGKEKNLEFTTSFGTCIPLRLRIQNKTDVKTDFIAEVSHSSIQIDKECCLGPLEKGKILVWFEPTQLGVQNCKVSFKSTLAGEFVFNIKGIATEPKPQGPYEVKAGGSTTIKFKNVFDSSRMFKIYVDREEFYVKTMFETIKPKKDLRISVYLNDKPAQGWTEVPTGCLTIETHEPAEPKVHWTYFLQGKQ
Summary
Description
Required for ciliary motility.
Keywords
Alternative splicing
Cell projection
Coiled coil
Complete proteome
Reference proteome
3D-structure
Ciliopathy
Kartagener syndrome
Polymorphism
Primary ciliary dyskinesia
Feature
chain Hydrocephalus-inducing protein
splice variant In isoform 3.
sequence variant In dbSNP:rs7200485.
splice variant In isoform 3.
sequence variant In dbSNP:rs7200485.
Uniprot
A0A2H1WKT1
A0A2A4JJ76
A0A212FA77
A0A194PP09
A0A194QSA7
A0A0L7LKW4
+ More
A0A2W1BSE4 A0A091CXW8 W5MHQ1 W5MHR6 V4BQE4 L9KKN1 F6WJJ0 A0A3P8IM41 H0WFN6 A0A2T7NHJ3 K1QL99 A0A0B7A3N7 A0A267FQF0 A0A183WAC8 A0A337SLB3 A0A1I8GWQ9 A0A1I8GKJ3 A0A315VMU0 A0A0B7A3D2 Q80W93 Q3MHU4 A0A232FAZ7 A0A0Q3WR32 G1PU02 Q5BJQ9 S4R9B3 A0A2C9K9X8 R7TDL7 A0A212DDA5 A0A3L7I1U8 A0A210QFE7 C3ZBW1 A0A093H3D0 S7P5L3 K7JQB3 L5MD55 A0A3Q0CV73 G5C1Q4 A0A091WNA9 A0A0A0AFS5 H2XZ27 U6DAW9 G3VZI4 A0A3Q0K1N7 A0A1L8GF23 G3T094 A0A1L8GF33 A0A1S3J5T2 A0A0F8CGF7 F6QBA5 A0A2K6GI20 G7Q1L1 A0A1S3EXT0 F1LSN6 A0A0G2JXD6 F6U0F6 W5P1K1 G1S2C7 G4VJV4 A0A2Y9E6A0 E1BED7 H2NRF5 A0A2J8VW78 A0A3Q0G7E4 L7MZG4 M7BNE7 A0A2U4BD20 A0A341AQ85 L5KUE2 A0A068XHE2 A0A1S3MN59 A0A1S3MN07 A0A1S3MMV0 G3URZ1 A0A2Y9SVU2 G1MRN0 A0A3P9MUW1 A0A3P9MV17 A0A146Y440 H2QBG9 K7G0R0 B1H3K7 Q4G0P3 G3RBF4 A0A2J8KA34 M3YFZ1 F1NYR0 H2YKD6 M3YFZ2 A0A1D5P4W4 I3K684 W5LN16 A0A2Y9K2D2 A0A1W4Z5Y2
A0A2W1BSE4 A0A091CXW8 W5MHQ1 W5MHR6 V4BQE4 L9KKN1 F6WJJ0 A0A3P8IM41 H0WFN6 A0A2T7NHJ3 K1QL99 A0A0B7A3N7 A0A267FQF0 A0A183WAC8 A0A337SLB3 A0A1I8GWQ9 A0A1I8GKJ3 A0A315VMU0 A0A0B7A3D2 Q80W93 Q3MHU4 A0A232FAZ7 A0A0Q3WR32 G1PU02 Q5BJQ9 S4R9B3 A0A2C9K9X8 R7TDL7 A0A212DDA5 A0A3L7I1U8 A0A210QFE7 C3ZBW1 A0A093H3D0 S7P5L3 K7JQB3 L5MD55 A0A3Q0CV73 G5C1Q4 A0A091WNA9 A0A0A0AFS5 H2XZ27 U6DAW9 G3VZI4 A0A3Q0K1N7 A0A1L8GF23 G3T094 A0A1L8GF33 A0A1S3J5T2 A0A0F8CGF7 F6QBA5 A0A2K6GI20 G7Q1L1 A0A1S3EXT0 F1LSN6 A0A0G2JXD6 F6U0F6 W5P1K1 G1S2C7 G4VJV4 A0A2Y9E6A0 E1BED7 H2NRF5 A0A2J8VW78 A0A3Q0G7E4 L7MZG4 M7BNE7 A0A2U4BD20 A0A341AQ85 L5KUE2 A0A068XHE2 A0A1S3MN59 A0A1S3MN07 A0A1S3MMV0 G3URZ1 A0A2Y9SVU2 G1MRN0 A0A3P9MUW1 A0A3P9MV17 A0A146Y440 H2QBG9 K7G0R0 B1H3K7 Q4G0P3 G3RBF4 A0A2J8KA34 M3YFZ1 F1NYR0 H2YKD6 M3YFZ2 A0A1D5P4W4 I3K684 W5LN16 A0A2Y9K2D2 A0A1W4Z5Y2
Pubmed
22118469
26354079
26227816
28756777
23254933
23385571
+ More
17495919 22992520 17975172 26392545 29703783 12719380 16141072 15489334 17296793 18250199 21183079 28648823 21993624 15562597 29294181 29704459 28812685 18563158 20075255 21993625 12481130 15114417 21709235 27762356 26383154 25835551 18464734 22002653 15057822 22673903 20809919 22253936 21881562 23624526 23258410 20838655 16136131 17381049 14702039 15616553 17974005 16938426 23022101 25186273 22398555 15592404 25186727 25329095
17495919 22992520 17975172 26392545 29703783 12719380 16141072 15489334 17296793 18250199 21183079 28648823 21993624 15562597 29294181 29704459 28812685 18563158 20075255 21993625 12481130 15114417 21709235 27762356 26383154 25835551 18464734 22002653 15057822 22673903 20809919 22253936 21881562 23624526 23258410 20838655 16136131 17381049 14702039 15616553 17974005 16938426 23022101 25186273 22398555 15592404 25186727 25329095
EMBL
ODYU01009244
SOQ53522.1
NWSH01001291
PCG71826.1
AGBW02009514
OWR50630.1
+ More
KQ459598 KPI94718.1 KQ461196 KPJ06426.1 JTDY01000796 KOB75846.1 KZ149933 PZC77251.1 KN123534 KFO24649.1 AHAT01020935 AHAT01020936 KB202284 ESO91099.1 KB320797 ELW63034.1 UZAO01062662 VDQ04961.1 AAQR03081674 PZQS01000012 PVD20612.1 JH816714 EKC34643.1 HACG01027710 CEK74575.1 NIVC01000845 PAA76040.1 AANG04002512 NHOQ01001396 PWA24892.1 HACG01027711 CEK74576.1 AY173049 AK030144 AK043971 AK142981 BB641870 BB664150 BC080316 BC104674 AAI04675.1 NNAY01000565 OXU27639.1 LMAW01000459 KQL50268.1 AAPE02023070 AAPE02023071 BC091376 AAH91376.1 AMQN01014785 KB311278 ELT89592.1 MKHE01000004 OWK16227.1 RAZU01000121 RLQ71611.1 NEDP02003910 OWF47429.1 GG666603 EEN50348.1 KL205731 KFV73572.1 KE161800 EPQ05523.1 KB101785 ELK36190.1 JH172853 EHB15465.1 KL411055 KFR03001.1 KL871350 KGL91870.1 HAAF01007423 CCP79247.1 AEFK01034944 AEFK01034945 AEFK01034946 AEFK01034947 AEFK01034948 AEFK01034949 AEFK01034950 AEFK01034951 AEFK01034952 AEFK01034953 CM004473 OCT82469.1 OCT82470.1 KQ040962 KKF32196.1 CM001295 EHH60549.1 AABR07043814 AABR07043815 AABR07043816 AABR07043817 CM001272 EHH31832.1 AMGL01028451 AMGL01028452 AMGL01028453 AMGL01028454 AMGL01028455 AMGL01028456 AMGL01028457 AMGL01028458 AMGL01028459 AMGL01028460 ADFV01127327 ADFV01127328 ADFV01127329 ADFV01127330 ADFV01127331 ADFV01127332 ADFV01127333 ADFV01127334 ADFV01127335 ADFV01127336 HE601627 CCD79705.1 ABGA01059428 ABGA01059429 ABGA01059430 ABGA01059431 ABGA01059432 ABGA01059433 ABGA01059434 ABGA01059435 ABGA01059436 ABGA01059437 NDHI03003408 PNJ61726.1 AAWZ02029921 KB554237 EMP29717.1 KB030572 ELK14581.1 GCES01037454 JAR48869.1 AACZ04050289 AACZ04050290 AACZ04050291 AACZ04050292 AACZ04050293 AACZ04050294 AACZ04050295 AGCU01130378 AGCU01130379 AGCU01130380 AGCU01130381 AGCU01130382 AGCU01130383 AGCU01130384 BC161431 AAI61431.1 AK022933 AK026688 AK299016 AK299348 AC027281 AC099495 AC138625 BC028351 BC043273 AL122038 AL133042 AL137259 CABD030101940 CABD030101941 CABD030101942 CABD030101943 CABD030101944 CABD030101945 CABD030101946 CABD030101947 CABD030101948 CABD030101949 NBAG03000381 PNI31888.1 AEYP01035514 AEYP01035515 AEYP01035516 AEYP01035517 AEYP01035518 AEYP01035519 AEYP01035520 AEYP01035521 AEYP01035522 AEYP01035523 AADN05000561 AERX01064686 AERX01064687 AERX01064688
KQ459598 KPI94718.1 KQ461196 KPJ06426.1 JTDY01000796 KOB75846.1 KZ149933 PZC77251.1 KN123534 KFO24649.1 AHAT01020935 AHAT01020936 KB202284 ESO91099.1 KB320797 ELW63034.1 UZAO01062662 VDQ04961.1 AAQR03081674 PZQS01000012 PVD20612.1 JH816714 EKC34643.1 HACG01027710 CEK74575.1 NIVC01000845 PAA76040.1 AANG04002512 NHOQ01001396 PWA24892.1 HACG01027711 CEK74576.1 AY173049 AK030144 AK043971 AK142981 BB641870 BB664150 BC080316 BC104674 AAI04675.1 NNAY01000565 OXU27639.1 LMAW01000459 KQL50268.1 AAPE02023070 AAPE02023071 BC091376 AAH91376.1 AMQN01014785 KB311278 ELT89592.1 MKHE01000004 OWK16227.1 RAZU01000121 RLQ71611.1 NEDP02003910 OWF47429.1 GG666603 EEN50348.1 KL205731 KFV73572.1 KE161800 EPQ05523.1 KB101785 ELK36190.1 JH172853 EHB15465.1 KL411055 KFR03001.1 KL871350 KGL91870.1 HAAF01007423 CCP79247.1 AEFK01034944 AEFK01034945 AEFK01034946 AEFK01034947 AEFK01034948 AEFK01034949 AEFK01034950 AEFK01034951 AEFK01034952 AEFK01034953 CM004473 OCT82469.1 OCT82470.1 KQ040962 KKF32196.1 CM001295 EHH60549.1 AABR07043814 AABR07043815 AABR07043816 AABR07043817 CM001272 EHH31832.1 AMGL01028451 AMGL01028452 AMGL01028453 AMGL01028454 AMGL01028455 AMGL01028456 AMGL01028457 AMGL01028458 AMGL01028459 AMGL01028460 ADFV01127327 ADFV01127328 ADFV01127329 ADFV01127330 ADFV01127331 ADFV01127332 ADFV01127333 ADFV01127334 ADFV01127335 ADFV01127336 HE601627 CCD79705.1 ABGA01059428 ABGA01059429 ABGA01059430 ABGA01059431 ABGA01059432 ABGA01059433 ABGA01059434 ABGA01059435 ABGA01059436 ABGA01059437 NDHI03003408 PNJ61726.1 AAWZ02029921 KB554237 EMP29717.1 KB030572 ELK14581.1 GCES01037454 JAR48869.1 AACZ04050289 AACZ04050290 AACZ04050291 AACZ04050292 AACZ04050293 AACZ04050294 AACZ04050295 AGCU01130378 AGCU01130379 AGCU01130380 AGCU01130381 AGCU01130382 AGCU01130383 AGCU01130384 BC161431 AAI61431.1 AK022933 AK026688 AK299016 AK299348 AC027281 AC099495 AC138625 BC028351 BC043273 AL122038 AL133042 AL137259 CABD030101940 CABD030101941 CABD030101942 CABD030101943 CABD030101944 CABD030101945 CABD030101946 CABD030101947 CABD030101948 CABD030101949 NBAG03000381 PNI31888.1 AEYP01035514 AEYP01035515 AEYP01035516 AEYP01035517 AEYP01035518 AEYP01035519 AEYP01035520 AEYP01035521 AEYP01035522 AEYP01035523 AADN05000561 AERX01064686 AERX01064687 AERX01064688
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
UP000028990
+ More
UP000018468 UP000030746 UP000011518 UP000002280 UP000005225 UP000245119 UP000005408 UP000215902 UP000050795 UP000011712 UP000095280 UP000000589 UP000215335 UP000051836 UP000001074 UP000245300 UP000076420 UP000014760 UP000273346 UP000242188 UP000001554 UP000053584 UP000002358 UP000189706 UP000006813 UP000053283 UP000053858 UP000008144 UP000007648 UP000186698 UP000007646 UP000085678 UP000002279 UP000233160 UP000009130 UP000081671 UP000002494 UP000002356 UP000001073 UP000008854 UP000248480 UP000001595 UP000189705 UP000001646 UP000031443 UP000245320 UP000252040 UP000010552 UP000087266 UP000001645 UP000248484 UP000242638 UP000002277 UP000007267 UP000005640 UP000001519 UP000000715 UP000000539 UP000007875 UP000005207 UP000018467 UP000248482 UP000192224
UP000018468 UP000030746 UP000011518 UP000002280 UP000005225 UP000245119 UP000005408 UP000215902 UP000050795 UP000011712 UP000095280 UP000000589 UP000215335 UP000051836 UP000001074 UP000245300 UP000076420 UP000014760 UP000273346 UP000242188 UP000001554 UP000053584 UP000002358 UP000189706 UP000006813 UP000053283 UP000053858 UP000008144 UP000007648 UP000186698 UP000007646 UP000085678 UP000002279 UP000233160 UP000009130 UP000081671 UP000002494 UP000002356 UP000001073 UP000008854 UP000248480 UP000001595 UP000189705 UP000001646 UP000031443 UP000245320 UP000252040 UP000010552 UP000087266 UP000001645 UP000248484 UP000242638 UP000002277 UP000007267 UP000005640 UP000001519 UP000000715 UP000000539 UP000007875 UP000005207 UP000018467 UP000248482 UP000192224
Pfam
Interpro
IPR013783
Ig-like_fold
+ More
IPR033305 Hydin-like
IPR008962 PapD-like_sf
IPR031549 ASH
IPR033768 Hydin_ADK
IPR027417 P-loop_NTPase
IPR023591 Ribosomal_S2_flav_dom_sf
IPR025807 Adrift-typ_MeTrfase
IPR029063 SAM-dependent_MTases
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR001955 Pancreatic_hormone-like
IPR000535 MSP_dom
IPR041322 SYCP2_ARLD
IPR002928 Myosin_tail
IPR000399 TPP-bd_CS
IPR033305 Hydin-like
IPR008962 PapD-like_sf
IPR031549 ASH
IPR033768 Hydin_ADK
IPR027417 P-loop_NTPase
IPR023591 Ribosomal_S2_flav_dom_sf
IPR025807 Adrift-typ_MeTrfase
IPR029063 SAM-dependent_MTases
IPR002877 rRNA_MeTrfase_FtsJ_dom
IPR001955 Pancreatic_hormone-like
IPR000535 MSP_dom
IPR041322 SYCP2_ARLD
IPR002928 Myosin_tail
IPR000399 TPP-bd_CS
Gene 3D
ProteinModelPortal
A0A2H1WKT1
A0A2A4JJ76
A0A212FA77
A0A194PP09
A0A194QSA7
A0A0L7LKW4
+ More
A0A2W1BSE4 A0A091CXW8 W5MHQ1 W5MHR6 V4BQE4 L9KKN1 F6WJJ0 A0A3P8IM41 H0WFN6 A0A2T7NHJ3 K1QL99 A0A0B7A3N7 A0A267FQF0 A0A183WAC8 A0A337SLB3 A0A1I8GWQ9 A0A1I8GKJ3 A0A315VMU0 A0A0B7A3D2 Q80W93 Q3MHU4 A0A232FAZ7 A0A0Q3WR32 G1PU02 Q5BJQ9 S4R9B3 A0A2C9K9X8 R7TDL7 A0A212DDA5 A0A3L7I1U8 A0A210QFE7 C3ZBW1 A0A093H3D0 S7P5L3 K7JQB3 L5MD55 A0A3Q0CV73 G5C1Q4 A0A091WNA9 A0A0A0AFS5 H2XZ27 U6DAW9 G3VZI4 A0A3Q0K1N7 A0A1L8GF23 G3T094 A0A1L8GF33 A0A1S3J5T2 A0A0F8CGF7 F6QBA5 A0A2K6GI20 G7Q1L1 A0A1S3EXT0 F1LSN6 A0A0G2JXD6 F6U0F6 W5P1K1 G1S2C7 G4VJV4 A0A2Y9E6A0 E1BED7 H2NRF5 A0A2J8VW78 A0A3Q0G7E4 L7MZG4 M7BNE7 A0A2U4BD20 A0A341AQ85 L5KUE2 A0A068XHE2 A0A1S3MN59 A0A1S3MN07 A0A1S3MMV0 G3URZ1 A0A2Y9SVU2 G1MRN0 A0A3P9MUW1 A0A3P9MV17 A0A146Y440 H2QBG9 K7G0R0 B1H3K7 Q4G0P3 G3RBF4 A0A2J8KA34 M3YFZ1 F1NYR0 H2YKD6 M3YFZ2 A0A1D5P4W4 I3K684 W5LN16 A0A2Y9K2D2 A0A1W4Z5Y2
A0A2W1BSE4 A0A091CXW8 W5MHQ1 W5MHR6 V4BQE4 L9KKN1 F6WJJ0 A0A3P8IM41 H0WFN6 A0A2T7NHJ3 K1QL99 A0A0B7A3N7 A0A267FQF0 A0A183WAC8 A0A337SLB3 A0A1I8GWQ9 A0A1I8GKJ3 A0A315VMU0 A0A0B7A3D2 Q80W93 Q3MHU4 A0A232FAZ7 A0A0Q3WR32 G1PU02 Q5BJQ9 S4R9B3 A0A2C9K9X8 R7TDL7 A0A212DDA5 A0A3L7I1U8 A0A210QFE7 C3ZBW1 A0A093H3D0 S7P5L3 K7JQB3 L5MD55 A0A3Q0CV73 G5C1Q4 A0A091WNA9 A0A0A0AFS5 H2XZ27 U6DAW9 G3VZI4 A0A3Q0K1N7 A0A1L8GF23 G3T094 A0A1L8GF33 A0A1S3J5T2 A0A0F8CGF7 F6QBA5 A0A2K6GI20 G7Q1L1 A0A1S3EXT0 F1LSN6 A0A0G2JXD6 F6U0F6 W5P1K1 G1S2C7 G4VJV4 A0A2Y9E6A0 E1BED7 H2NRF5 A0A2J8VW78 A0A3Q0G7E4 L7MZG4 M7BNE7 A0A2U4BD20 A0A341AQ85 L5KUE2 A0A068XHE2 A0A1S3MN59 A0A1S3MN07 A0A1S3MMV0 G3URZ1 A0A2Y9SVU2 G1MRN0 A0A3P9MUW1 A0A3P9MV17 A0A146Y440 H2QBG9 K7G0R0 B1H3K7 Q4G0P3 G3RBF4 A0A2J8KA34 M3YFZ1 F1NYR0 H2YKD6 M3YFZ2 A0A1D5P4W4 I3K684 W5LN16 A0A2Y9K2D2 A0A1W4Z5Y2
Ontologies
KEGG
GO
PANTHER
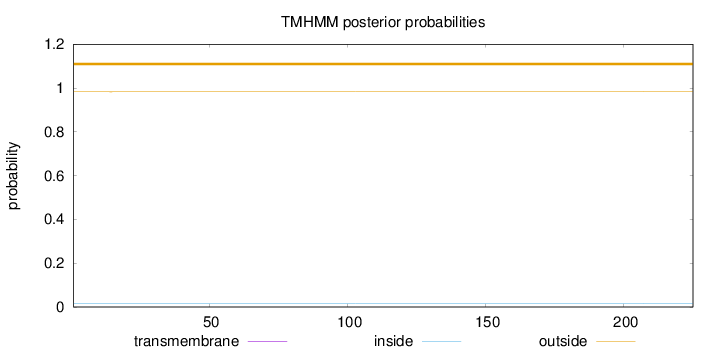
Topology
Subcellular location
Length:
225
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00285
Exp number, first 60 AAs:
0.00285
Total prob of N-in:
0.01722
outside
1 - 225
Population Genetic Test Statistics
Pi
115.662874
Theta
128.747556
Tajima's D
-0.40091
CLR
2.065201
CSRT
0.266786660666967
Interpretation
Uncertain
