Pre Gene Modal
BGIBMGA003165
Annotation
PREDICTED:_transmembrane_BAX_inhibitor_motif-containing_protein_5_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.667
Sequence
CDS
ATGTGTATCGGTCGCTCGGCGTTCAATGTAACGCAGAGTCTAAAATCGCCAGTACCACAAAATTTTGTACCGAGAAACTATGTGGTCAGAAATTATGCACGAGAACCTCGAACAAGGGTCGCTACACGCTCACAACCCACACTTCGGGAGAGACTCATGGCACCTGCTGGACCAAATGCATTCAGCTTGGGCAAGGGTGCATTAGCTGGAGCCTCTGCAGTTGGACTAGTTGCTCTGTGCTACTATGGCTCTGGGGTAAAACCAGGTACTCTTCAGGAATCTCACCTTTGGCCACAATATGTGAAAGAACGTATAAAAACAACATATGGATACATCGCGGGTTCTTTGGTGCTGACTGCTGGTAGTGCAGTTGCTGTTTTCAGAACACCCGCACTTCTAAATTTGGTAGCAAGAAATGGATGGATGTCCATTATTGTCACATTGGGTCTGATGATTGGTTCTGGCATGGTAGTAAGAGGAATGGAGTACACACCTGGATTTGGGGCTAAACAATTAGCATGGATGGCGCACACTGGTATTATGGGAGCTGTCATTGCTCCAATTTGCTTCCTTGGTGGCCCAATCCTGATGCGTGCAGCTTGGTACACTGCTGGTGTAGTTGGTGGTCTGAGTACAATTGCAGTGTGTGCACCATCTGGAGAGTTCTTGAACATGCGTGCTCCACTTGCTATGGGTCTAGGAGCTGTGTTTGCTGCATCTCTGGCTGGGATGTTTCTACCACCTACTAGTGCTCTTGGTGCCGGATTGTATTCACTAAGTTTGTATGGTGGCTTAATTGTATTTGGCGGATTTTTATTATACGATACGCAATCTATTATTAAACGCGCCGAGATGCATCCACCCTACGGATTCAAACCATATGACCCAATCAATTCGGCCATATCTGTTTATCTGGATGTTCTGAATATATTCATGAGAATTGCCATGATCTTGTCTGGAGCTGGTGGCAACAGGAGGAAGTAA
Protein
MCIGRSAFNVTQSLKSPVPQNFVPRNYVVRNYAREPRTRVATRSQPTLRERLMAPAGPNAFSLGKGALAGASAVGLVALCYYGSGVKPGTLQESHLWPQYVKERIKTTYGYIAGSLVLTAGSAVAVFRTPALLNLVARNGWMSIIVTLGLMIGSGMVVRGMEYTPGFGAKQLAWMAHTGIMGAVIAPICFLGGPILMRAAWYTAGVVGGLSTIAVCAPSGEFLNMRAPLAMGLGAVFAASLAGMFLPPTSALGAGLYSLSLYGGLIVFGGFLLYDTQSIIKRAEMHPPYGFKPYDPINSAISVYLDVLNIFMRIAMILSGAGGNRRK
Summary
Similarity
Belongs to the BI1 family.
Uniprot
Q2F5I3
Q283L9
S4NXE1
A0A212FA70
A0A194PUA6
A0A1E1WN82
+ More
A0A2W1BZK0 A0A2A4JJC8 A0A194QLJ0 A0A2H1X165 H9J0X8 A0A1Q3FKX5 A0A0P6IV18 B0WCS4 U5EQS8 T1E2S4 A0A182IUW2 A0A1J1HPZ6 A0A0K8TSG9 A0A2R5L6A8 A0A182WGV1 A0A293LKD6 A0A182K3S1 A0A2M4BU35 A0A2M4BT65 A0A0L0C246 A0A2M4AAF2 A0A1S3HCW4 A0A182Q0T1 W5JBS2 T1E8A5 A0A1Y1L0P1 A0A182PJI4 A0A087TM87 A0A0K8REW7 A0A1L8EEQ8 A0A1Z5L5X0 A0A023ER86 A0A131Y495 T1PER1 A0A1B6DYK6 T1E2T4 W8C0K3 A0A0K8W7P3 A0A0A1WG24 A0A1E1XHB3 A0A182NUI7 A0A182RAS4 A0A182F2I2 A0A182LZQ7 A0A023GIS5 G3MPQ7 Q5TP14 A0A182YDN3 A0A182VIV2 A0A182KTM3 A0A182HRK0 A0A084WC79 A0A1B0CR00 A0A182TH31 A0A224YYC2 A0A023FZ01 A0A131YRT9 A0A131XPE6 A0A023FIG4 A0A1I8NXH5 D6WNY7 B4N890 A0A023FKD5 A0A2R7WHP2 A0A1A9WIN4 A0A3B0K6V5 A0A1L8DK15 B5DM07 B4H4S1 A0A0C9R245 E0VC33 A0A164KRT9 A0A0P5F8W3 A0A0T6AU37 A0A1B0D2M2 A0A1V9XH30 B4MAM0 B3MQ84 B4I4U6 B4JF90 A0A232FEA6 B4QZN4 Q9VIB2 K7IP50 B3P2X8 A0A1W4V2X3 V9IAT0 A0A1W4VN03 B4PRH5 E9H821 Q8T8Z4 B4L712 A0A069DYN7
A0A2W1BZK0 A0A2A4JJC8 A0A194QLJ0 A0A2H1X165 H9J0X8 A0A1Q3FKX5 A0A0P6IV18 B0WCS4 U5EQS8 T1E2S4 A0A182IUW2 A0A1J1HPZ6 A0A0K8TSG9 A0A2R5L6A8 A0A182WGV1 A0A293LKD6 A0A182K3S1 A0A2M4BU35 A0A2M4BT65 A0A0L0C246 A0A2M4AAF2 A0A1S3HCW4 A0A182Q0T1 W5JBS2 T1E8A5 A0A1Y1L0P1 A0A182PJI4 A0A087TM87 A0A0K8REW7 A0A1L8EEQ8 A0A1Z5L5X0 A0A023ER86 A0A131Y495 T1PER1 A0A1B6DYK6 T1E2T4 W8C0K3 A0A0K8W7P3 A0A0A1WG24 A0A1E1XHB3 A0A182NUI7 A0A182RAS4 A0A182F2I2 A0A182LZQ7 A0A023GIS5 G3MPQ7 Q5TP14 A0A182YDN3 A0A182VIV2 A0A182KTM3 A0A182HRK0 A0A084WC79 A0A1B0CR00 A0A182TH31 A0A224YYC2 A0A023FZ01 A0A131YRT9 A0A131XPE6 A0A023FIG4 A0A1I8NXH5 D6WNY7 B4N890 A0A023FKD5 A0A2R7WHP2 A0A1A9WIN4 A0A3B0K6V5 A0A1L8DK15 B5DM07 B4H4S1 A0A0C9R245 E0VC33 A0A164KRT9 A0A0P5F8W3 A0A0T6AU37 A0A1B0D2M2 A0A1V9XH30 B4MAM0 B3MQ84 B4I4U6 B4JF90 A0A232FEA6 B4QZN4 Q9VIB2 K7IP50 B3P2X8 A0A1W4V2X3 V9IAT0 A0A1W4VN03 B4PRH5 E9H821 Q8T8Z4 B4L712 A0A069DYN7
Pubmed
16495681
23622113
22118469
26354079
28756777
19121390
+ More
26999592 24330624 26369729 26108605 20920257 23761445 28004739 28528879 24945155 26483478 25315136 24495485 25830018 28503490 22216098 12364791 14747013 17210077 25244985 20966253 24438588 28797301 26830274 28049606 18362917 19820115 17994087 15632085 26131772 20566863 28327890 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 17550304 21292972 18057021 26334808
26999592 24330624 26369729 26108605 20920257 23761445 28004739 28528879 24945155 26483478 25315136 24495485 25830018 28503490 22216098 12364791 14747013 17210077 25244985 20966253 24438588 28797301 26830274 28049606 18362917 19820115 17994087 15632085 26131772 20566863 28327890 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20075255 17550304 21292972 18057021 26334808
EMBL
DQ311440
ABD36384.1
DQ228724
ABB49054.1
GAIX01014220
JAA78340.1
+ More
AGBW02009514 OWR50632.1 KQ459598 KPI94720.1 GDQN01003490 GDQN01002612 JAT87564.1 JAT88442.1 KZ149933 PZC77253.1 NWSH01001291 PCG71828.1 KQ461196 KPJ06428.1 ODYU01012632 SOQ59051.1 BABH01010446 BABH01010447 GFDL01006806 JAV28239.1 GDUN01000347 JAN95572.1 DS231890 EDS43800.1 GANO01003145 JAB56726.1 GALA01001035 JAA93817.1 CVRI01000013 CRK89602.1 GDAI01000723 JAI16880.1 GGLE01000902 MBY05028.1 GFWV01004453 MAA29183.1 GGFJ01007127 MBW56268.1 GGFJ01007126 MBW56267.1 JRES01000994 KNC26341.1 GGFK01004445 MBW37766.1 AXCN02002061 ADMH02001853 ETN60863.1 GAMD01002943 JAA98647.1 GEZM01068062 GEZM01068061 JAV67239.1 KK115857 KFM66226.1 GADI01004385 JAA69423.1 GFDG01001664 JAV17135.1 GFJQ02004221 JAW02749.1 JXUM01103896 GAPW01002435 GAPW01002434 KQ564842 JAC11164.1 KXJ71663.1 GEFM01003055 JAP72741.1 KA646630 AFP61259.1 GEDC01014654 GEDC01006544 JAS22644.1 JAS30754.1 GALA01001010 JAA93842.1 GAMC01001353 GAMC01001352 JAC05203.1 GDHF01006619 GDHF01005245 JAI45695.1 JAI47069.1 GBXI01016495 JAC97796.1 GFAC01000481 JAT98707.1 AXCM01000820 GBBM01001627 JAC33791.1 JO843858 AEO35475.1 AAAB01008980 EAL39226.3 APCN01001715 ATLV01022579 ATLV01022580 KE525333 KFB47823.1 AJWK01024261 GFPF01008177 MAA19323.1 GBBL01001395 JAC25925.1 GEDV01007302 JAP81255.1 GEFH01000563 JAP68018.1 GBBK01002881 JAC21601.1 KQ971343 EFA03190.1 CH964232 EDW81341.1 GBBK01002883 JAC21599.1 KK854831 PTY19088.1 OUUW01000035 SPP89844.1 GFDF01007308 JAV06776.1 CH379064 EDY72523.1 CH479209 EDW32683.1 GBZX01001655 JAG91085.1 DS235045 EEB10939.1 LRGB01003257 KZS03483.1 GDIQ01258423 JAJ93301.1 LJIG01022848 KRT78405.1 AJVK01010693 AJVK01010694 MNPL01011269 OQR72672.1 CH940655 EDW66279.1 CH902621 EDV44510.1 CH480821 EDW55239.1 CH916369 EDV93371.1 NNAY01000372 OXU28818.1 CM000364 EDX11992.1 AE014297 BT082005 AAF54013.2 ACO57624.1 CH954181 EDV48150.1 JR038380 JR038381 AEY58227.1 AEY58228.1 CM000160 EDW96363.1 GL732602 EFX72148.1 AY075204 AAL68072.1 CH933812 EDW06158.1 KRG07164.1 GBGD01002070 JAC86819.1
AGBW02009514 OWR50632.1 KQ459598 KPI94720.1 GDQN01003490 GDQN01002612 JAT87564.1 JAT88442.1 KZ149933 PZC77253.1 NWSH01001291 PCG71828.1 KQ461196 KPJ06428.1 ODYU01012632 SOQ59051.1 BABH01010446 BABH01010447 GFDL01006806 JAV28239.1 GDUN01000347 JAN95572.1 DS231890 EDS43800.1 GANO01003145 JAB56726.1 GALA01001035 JAA93817.1 CVRI01000013 CRK89602.1 GDAI01000723 JAI16880.1 GGLE01000902 MBY05028.1 GFWV01004453 MAA29183.1 GGFJ01007127 MBW56268.1 GGFJ01007126 MBW56267.1 JRES01000994 KNC26341.1 GGFK01004445 MBW37766.1 AXCN02002061 ADMH02001853 ETN60863.1 GAMD01002943 JAA98647.1 GEZM01068062 GEZM01068061 JAV67239.1 KK115857 KFM66226.1 GADI01004385 JAA69423.1 GFDG01001664 JAV17135.1 GFJQ02004221 JAW02749.1 JXUM01103896 GAPW01002435 GAPW01002434 KQ564842 JAC11164.1 KXJ71663.1 GEFM01003055 JAP72741.1 KA646630 AFP61259.1 GEDC01014654 GEDC01006544 JAS22644.1 JAS30754.1 GALA01001010 JAA93842.1 GAMC01001353 GAMC01001352 JAC05203.1 GDHF01006619 GDHF01005245 JAI45695.1 JAI47069.1 GBXI01016495 JAC97796.1 GFAC01000481 JAT98707.1 AXCM01000820 GBBM01001627 JAC33791.1 JO843858 AEO35475.1 AAAB01008980 EAL39226.3 APCN01001715 ATLV01022579 ATLV01022580 KE525333 KFB47823.1 AJWK01024261 GFPF01008177 MAA19323.1 GBBL01001395 JAC25925.1 GEDV01007302 JAP81255.1 GEFH01000563 JAP68018.1 GBBK01002881 JAC21601.1 KQ971343 EFA03190.1 CH964232 EDW81341.1 GBBK01002883 JAC21599.1 KK854831 PTY19088.1 OUUW01000035 SPP89844.1 GFDF01007308 JAV06776.1 CH379064 EDY72523.1 CH479209 EDW32683.1 GBZX01001655 JAG91085.1 DS235045 EEB10939.1 LRGB01003257 KZS03483.1 GDIQ01258423 JAJ93301.1 LJIG01022848 KRT78405.1 AJVK01010693 AJVK01010694 MNPL01011269 OQR72672.1 CH940655 EDW66279.1 CH902621 EDV44510.1 CH480821 EDW55239.1 CH916369 EDV93371.1 NNAY01000372 OXU28818.1 CM000364 EDX11992.1 AE014297 BT082005 AAF54013.2 ACO57624.1 CH954181 EDV48150.1 JR038380 JR038381 AEY58227.1 AEY58228.1 CM000160 EDW96363.1 GL732602 EFX72148.1 AY075204 AAL68072.1 CH933812 EDW06158.1 KRG07164.1 GBGD01002070 JAC86819.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000053240
UP000005204
UP000002320
+ More
UP000075880 UP000183832 UP000075920 UP000075881 UP000037069 UP000085678 UP000075886 UP000000673 UP000075885 UP000054359 UP000069940 UP000249989 UP000095301 UP000075884 UP000075900 UP000069272 UP000075883 UP000007062 UP000076408 UP000075903 UP000075882 UP000075840 UP000030765 UP000092461 UP000075902 UP000095300 UP000007266 UP000007798 UP000091820 UP000268350 UP000001819 UP000008744 UP000009046 UP000076858 UP000092462 UP000192247 UP000008792 UP000007801 UP000001292 UP000001070 UP000215335 UP000000304 UP000000803 UP000002358 UP000008711 UP000192221 UP000002282 UP000000305 UP000009192
UP000075880 UP000183832 UP000075920 UP000075881 UP000037069 UP000085678 UP000075886 UP000000673 UP000075885 UP000054359 UP000069940 UP000249989 UP000095301 UP000075884 UP000075900 UP000069272 UP000075883 UP000007062 UP000076408 UP000075903 UP000075882 UP000075840 UP000030765 UP000092461 UP000075902 UP000095300 UP000007266 UP000007798 UP000091820 UP000268350 UP000001819 UP000008744 UP000009046 UP000076858 UP000092462 UP000192247 UP000008792 UP000007801 UP000001292 UP000001070 UP000215335 UP000000304 UP000000803 UP000002358 UP000008711 UP000192221 UP000002282 UP000000305 UP000009192
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
Q2F5I3
Q283L9
S4NXE1
A0A212FA70
A0A194PUA6
A0A1E1WN82
+ More
A0A2W1BZK0 A0A2A4JJC8 A0A194QLJ0 A0A2H1X165 H9J0X8 A0A1Q3FKX5 A0A0P6IV18 B0WCS4 U5EQS8 T1E2S4 A0A182IUW2 A0A1J1HPZ6 A0A0K8TSG9 A0A2R5L6A8 A0A182WGV1 A0A293LKD6 A0A182K3S1 A0A2M4BU35 A0A2M4BT65 A0A0L0C246 A0A2M4AAF2 A0A1S3HCW4 A0A182Q0T1 W5JBS2 T1E8A5 A0A1Y1L0P1 A0A182PJI4 A0A087TM87 A0A0K8REW7 A0A1L8EEQ8 A0A1Z5L5X0 A0A023ER86 A0A131Y495 T1PER1 A0A1B6DYK6 T1E2T4 W8C0K3 A0A0K8W7P3 A0A0A1WG24 A0A1E1XHB3 A0A182NUI7 A0A182RAS4 A0A182F2I2 A0A182LZQ7 A0A023GIS5 G3MPQ7 Q5TP14 A0A182YDN3 A0A182VIV2 A0A182KTM3 A0A182HRK0 A0A084WC79 A0A1B0CR00 A0A182TH31 A0A224YYC2 A0A023FZ01 A0A131YRT9 A0A131XPE6 A0A023FIG4 A0A1I8NXH5 D6WNY7 B4N890 A0A023FKD5 A0A2R7WHP2 A0A1A9WIN4 A0A3B0K6V5 A0A1L8DK15 B5DM07 B4H4S1 A0A0C9R245 E0VC33 A0A164KRT9 A0A0P5F8W3 A0A0T6AU37 A0A1B0D2M2 A0A1V9XH30 B4MAM0 B3MQ84 B4I4U6 B4JF90 A0A232FEA6 B4QZN4 Q9VIB2 K7IP50 B3P2X8 A0A1W4V2X3 V9IAT0 A0A1W4VN03 B4PRH5 E9H821 Q8T8Z4 B4L712 A0A069DYN7
A0A2W1BZK0 A0A2A4JJC8 A0A194QLJ0 A0A2H1X165 H9J0X8 A0A1Q3FKX5 A0A0P6IV18 B0WCS4 U5EQS8 T1E2S4 A0A182IUW2 A0A1J1HPZ6 A0A0K8TSG9 A0A2R5L6A8 A0A182WGV1 A0A293LKD6 A0A182K3S1 A0A2M4BU35 A0A2M4BT65 A0A0L0C246 A0A2M4AAF2 A0A1S3HCW4 A0A182Q0T1 W5JBS2 T1E8A5 A0A1Y1L0P1 A0A182PJI4 A0A087TM87 A0A0K8REW7 A0A1L8EEQ8 A0A1Z5L5X0 A0A023ER86 A0A131Y495 T1PER1 A0A1B6DYK6 T1E2T4 W8C0K3 A0A0K8W7P3 A0A0A1WG24 A0A1E1XHB3 A0A182NUI7 A0A182RAS4 A0A182F2I2 A0A182LZQ7 A0A023GIS5 G3MPQ7 Q5TP14 A0A182YDN3 A0A182VIV2 A0A182KTM3 A0A182HRK0 A0A084WC79 A0A1B0CR00 A0A182TH31 A0A224YYC2 A0A023FZ01 A0A131YRT9 A0A131XPE6 A0A023FIG4 A0A1I8NXH5 D6WNY7 B4N890 A0A023FKD5 A0A2R7WHP2 A0A1A9WIN4 A0A3B0K6V5 A0A1L8DK15 B5DM07 B4H4S1 A0A0C9R245 E0VC33 A0A164KRT9 A0A0P5F8W3 A0A0T6AU37 A0A1B0D2M2 A0A1V9XH30 B4MAM0 B3MQ84 B4I4U6 B4JF90 A0A232FEA6 B4QZN4 Q9VIB2 K7IP50 B3P2X8 A0A1W4V2X3 V9IAT0 A0A1W4VN03 B4PRH5 E9H821 Q8T8Z4 B4L712 A0A069DYN7
Ontologies
PANTHER
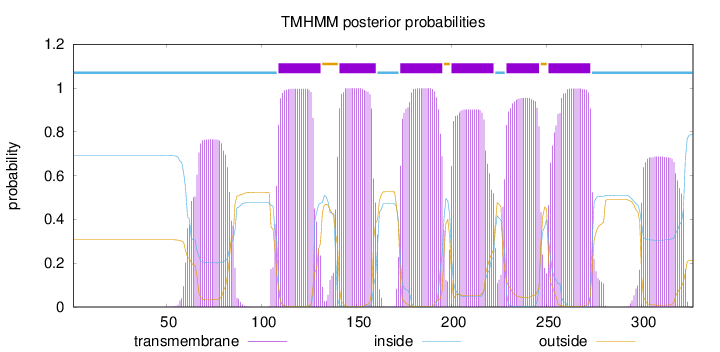
Topology
Length:
327
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
157.30979
Exp number, first 60 AAs:
0.38918
Total prob of N-in:
0.69273
inside
1 - 108
TMhelix
109 - 131
outside
132 - 140
TMhelix
141 - 160
inside
161 - 172
TMhelix
173 - 195
outside
196 - 199
TMhelix
200 - 222
inside
223 - 228
TMhelix
229 - 246
outside
247 - 250
TMhelix
251 - 273
inside
274 - 327
Population Genetic Test Statistics
Pi
199.466724
Theta
221.312425
Tajima's D
-0.424602
CLR
2.058976
CSRT
0.260336983150842
Interpretation
Possibly Positive selection
