Gene
KWMTBOMO02118
Pre Gene Modal
BGIBMGA002999
Annotation
hypothetical_protein_KGM_01625_[Danaus_plexippus]
Full name
Tetraspanin
Location in the cell
PlasmaMembrane Reliability : 4.239
Sequence
CDS
ATGGGAAATCAATTTAAGAACGTGGCGGTCATCGCCTGCTTGAAGACTTTTCTTTTCGTTTTCAATGTTGTGTTTTGGTTAACTGGGCTACTACTGTTGGTGGTAGGACTATGGGCAGAGTTCGATCTACATAAATACCTGGAACTCTCACCGGAGTTCTCCGGGACAGCTCCTCATGTCATGATTGGAATCGCCGGCTTAATCGTACTGATCAGTTCCGTGGCCTTTTCCTGTATCATCAAAGGACAACCCGTCTTGCTTTACATCTACGGTGGTTTCTTGGCATGCATATTTATGATGGAAGCCGGCGTTGGAGCTTCTGTAGCATGTTACCGCGACACCTTCGCTAAGGGTCTTCGCGACGGACTTACCGAAACAATTACTCACTACAACCCACAGAAAGCAAACTTTGATTTCGCGCAAACCAAGTTGCATTGCTGTGGCGTAAATAACTACACGGACTGGATGAGGATGTCGCCACAGCGCGTGATCCCTATCTCCTGCTGCGTAGACGCTAATAACTGCGTCACGGCCAATTACAGCGAAGTCCATCAAAAGACAAAAATAAACATAGTAAAAGATAGCTTCACTTTCGAATGA
Protein
MGNQFKNVAVIACLKTFLFVFNVVFWLTGLLLLVVGLWAEFDLHKYLELSPEFSGTAPHVMIGIAGLIVLISSVAFSCIIKGQPVLLYIYGGFLACIFMMEAGVGASVACYRDTFAKGLRDGLTETITHYNPQKANFDFAQTKLHCCGVNNYTDWMRMSPQRVIPISCCVDANNCVTANYSEVHQKTKINIVKDSFTFE
Summary
Similarity
Belongs to the tetraspanin (TM4SF) family.
Uniprot
H9J0G2
A0A212FA61
A0A2A4JK78
A0A2H1V280
A0A2W1BX49
A0A194PP14
+ More
A0A194QSB2 A0A0L7L883 D6WNZ1 A0A2J7RAC0 A0A1Y1LWZ5 A0A1B0D099 U4TY90 U5EVC2 A0A0T6BC03 A0A2P8XHC9 K7IRI9 A0A087UPS1 A0A1D2NHQ3 A0A2L2YQI9 T1IN95 A0A1J1IQH4 A0A2P6KYT6 A0A2T7NC29 E9IK86 E2BKP2 A0A0C9QX11 V3ZNB1 C3YK57 A0A088ABB0 K1Q8A5 A0A2A3E0X8 V9ICQ6 Q2TCS1 A0A0M9ABC1 A0A0L7QW36 A0A151XG20 A0A195CTI3 A0A151IXH2 F4WYM6 A0A2C9LYG3 A0A210QYA5 A0A3P9BIC1 A0A3P8N992 A0A3B4FXW5 A0A2R5LE09 A0A195BY00 A0A154P8E8 A0A0L8I669 E2AFI6 A0A0L0CFQ3 T1FN76 N6TLA3 A0A195F0D5 A0A146X6Y6 A0A1S3JDE5 R7UTH9 A0A146X895 A0A0C9RC81 A0A034WAS6 I3J346 A0A060X046 A0A3B3VI42 A0A147ACX0 A0A3B1JWB4 A0A3B4X585 A0A3B4T5K5 A0A1A8MND5 A0A3P8SA18 I3JHY6 A0A3B3DL88 A0A060WYB6 A0A3B4CRH3 A0A1A8G6E6 A0A3B4G2L7 A0A3P8RGM8 A0A3P9D4U8 A0A1A8B5H4 A0A3B3DL83 K4GIK4 E9FX18 A0A146MK15 A0A1S3MSP3 A0A1S3RSG3 K4FUU5 H2M7Y3 A0A3P9M7E8 A0A3P9I500 A0A2U9C8B9 A0A0C9Q8W8 A0A3B3DJ63 A0A3B3IC82 V9KQW5 A0A3P9I5F1 A0A3B3I5I1 H2L751
A0A194QSB2 A0A0L7L883 D6WNZ1 A0A2J7RAC0 A0A1Y1LWZ5 A0A1B0D099 U4TY90 U5EVC2 A0A0T6BC03 A0A2P8XHC9 K7IRI9 A0A087UPS1 A0A1D2NHQ3 A0A2L2YQI9 T1IN95 A0A1J1IQH4 A0A2P6KYT6 A0A2T7NC29 E9IK86 E2BKP2 A0A0C9QX11 V3ZNB1 C3YK57 A0A088ABB0 K1Q8A5 A0A2A3E0X8 V9ICQ6 Q2TCS1 A0A0M9ABC1 A0A0L7QW36 A0A151XG20 A0A195CTI3 A0A151IXH2 F4WYM6 A0A2C9LYG3 A0A210QYA5 A0A3P9BIC1 A0A3P8N992 A0A3B4FXW5 A0A2R5LE09 A0A195BY00 A0A154P8E8 A0A0L8I669 E2AFI6 A0A0L0CFQ3 T1FN76 N6TLA3 A0A195F0D5 A0A146X6Y6 A0A1S3JDE5 R7UTH9 A0A146X895 A0A0C9RC81 A0A034WAS6 I3J346 A0A060X046 A0A3B3VI42 A0A147ACX0 A0A3B1JWB4 A0A3B4X585 A0A3B4T5K5 A0A1A8MND5 A0A3P8SA18 I3JHY6 A0A3B3DL88 A0A060WYB6 A0A3B4CRH3 A0A1A8G6E6 A0A3B4G2L7 A0A3P8RGM8 A0A3P9D4U8 A0A1A8B5H4 A0A3B3DL83 K4GIK4 E9FX18 A0A146MK15 A0A1S3MSP3 A0A1S3RSG3 K4FUU5 H2M7Y3 A0A3P9M7E8 A0A3P9I500 A0A2U9C8B9 A0A0C9Q8W8 A0A3B3DJ63 A0A3B3IC82 V9KQW5 A0A3P9I5F1 A0A3B3I5I1 H2L751
Pubmed
EMBL
BABH01010450
BABH01010451
AGBW02009514
OWR50635.1
NWSH01001291
PCG71833.1
+ More
ODYU01000329 SOQ34876.1 KZ149933 PZC77256.1 KQ459598 KPI94723.1 KQ461196 KPJ06431.1 JTDY01002298 KOB71712.1 KQ971343 EFA04407.1 NEVH01006574 PNF37775.1 GEZM01049034 GEZM01049033 JAV76156.1 AJVK01002291 KB630971 KB631830 KB632086 ERL83974.1 ERL86589.1 ERL88647.1 GANO01001942 JAB57929.1 LJIG01002080 KRT84860.1 PYGN01002128 PSN31396.1 KK120917 KFM79360.1 LJIJ01000036 ODN04781.1 IAAA01045326 LAA10366.1 JH431152 CVRI01000055 CRL01350.1 MWRG01003527 PRD31487.1 PZQS01000014 PVD18718.1 GL763924 EFZ19049.1 GL448819 EFN83740.1 GBYB01005217 JAG74984.1 KB202014 ESO92848.1 GG666520 EEN59514.1 JH816697 EKC25090.1 KZ288469 PBC25367.1 JR038403 AEY58241.1 AY955263 AAY53605.1 KQ435692 KOX81047.1 KQ414716 KOC62827.1 KQ982174 KYQ59342.1 KQ977304 KYN03802.1 KQ980809 KYN12724.1 GL888453 EGI60714.1 NEDP02001253 OWF53682.1 GGLE01003644 MBY07770.1 KQ976394 KYM93170.1 KQ434844 KZC08195.1 KQ416546 KOF96520.1 GL439108 EFN67879.1 JRES01000453 KNC31071.1 AMQM01005151 KB096830 ESO01092.1 APGK01032329 APGK01032330 APGK01032331 KB740839 ENN78758.1 KQ981880 KYN33938.1 GCES01048445 JAR37878.1 AMQN01000981 KB298217 ELU09463.1 GCES01048444 JAR37879.1 GBYB01010717 JAG80484.1 GAKP01008044 GAKP01008043 JAC50908.1 AERX01013175 FR904845 CDQ72622.1 GCES01010321 JAR76002.1 HAEF01016775 SBR57934.1 AERX01026718 FR904698 CDQ70069.1 HAEB01019412 SBQ65939.1 HADY01023451 SBP61936.1 JX212614 AFM90928.1 GL732526 EFX87992.1 GCES01166767 JAQ19555.1 JX053146 AFK11374.1 CP026255 AWP11869.1 GBYB01010718 JAG80485.1 JW868218 AFP00736.1
ODYU01000329 SOQ34876.1 KZ149933 PZC77256.1 KQ459598 KPI94723.1 KQ461196 KPJ06431.1 JTDY01002298 KOB71712.1 KQ971343 EFA04407.1 NEVH01006574 PNF37775.1 GEZM01049034 GEZM01049033 JAV76156.1 AJVK01002291 KB630971 KB631830 KB632086 ERL83974.1 ERL86589.1 ERL88647.1 GANO01001942 JAB57929.1 LJIG01002080 KRT84860.1 PYGN01002128 PSN31396.1 KK120917 KFM79360.1 LJIJ01000036 ODN04781.1 IAAA01045326 LAA10366.1 JH431152 CVRI01000055 CRL01350.1 MWRG01003527 PRD31487.1 PZQS01000014 PVD18718.1 GL763924 EFZ19049.1 GL448819 EFN83740.1 GBYB01005217 JAG74984.1 KB202014 ESO92848.1 GG666520 EEN59514.1 JH816697 EKC25090.1 KZ288469 PBC25367.1 JR038403 AEY58241.1 AY955263 AAY53605.1 KQ435692 KOX81047.1 KQ414716 KOC62827.1 KQ982174 KYQ59342.1 KQ977304 KYN03802.1 KQ980809 KYN12724.1 GL888453 EGI60714.1 NEDP02001253 OWF53682.1 GGLE01003644 MBY07770.1 KQ976394 KYM93170.1 KQ434844 KZC08195.1 KQ416546 KOF96520.1 GL439108 EFN67879.1 JRES01000453 KNC31071.1 AMQM01005151 KB096830 ESO01092.1 APGK01032329 APGK01032330 APGK01032331 KB740839 ENN78758.1 KQ981880 KYN33938.1 GCES01048445 JAR37878.1 AMQN01000981 KB298217 ELU09463.1 GCES01048444 JAR37879.1 GBYB01010717 JAG80484.1 GAKP01008044 GAKP01008043 JAC50908.1 AERX01013175 FR904845 CDQ72622.1 GCES01010321 JAR76002.1 HAEF01016775 SBR57934.1 AERX01026718 FR904698 CDQ70069.1 HAEB01019412 SBQ65939.1 HADY01023451 SBP61936.1 JX212614 AFM90928.1 GL732526 EFX87992.1 GCES01166767 JAQ19555.1 JX053146 AFK11374.1 CP026255 AWP11869.1 GBYB01010718 JAG80485.1 JW868218 AFP00736.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
+ More
UP000007266 UP000235965 UP000092462 UP000030742 UP000245037 UP000002358 UP000054359 UP000094527 UP000183832 UP000245119 UP000008237 UP000030746 UP000001554 UP000005203 UP000005408 UP000242457 UP000053105 UP000053825 UP000075809 UP000078542 UP000078492 UP000007755 UP000076420 UP000242188 UP000265160 UP000265100 UP000261460 UP000078540 UP000076502 UP000053454 UP000000311 UP000037069 UP000015101 UP000019118 UP000078541 UP000085678 UP000014760 UP000005207 UP000193380 UP000261500 UP000018467 UP000261360 UP000261420 UP000265080 UP000261560 UP000261440 UP000000305 UP000087266 UP000001038 UP000265180 UP000265200 UP000246464
UP000007266 UP000235965 UP000092462 UP000030742 UP000245037 UP000002358 UP000054359 UP000094527 UP000183832 UP000245119 UP000008237 UP000030746 UP000001554 UP000005203 UP000005408 UP000242457 UP000053105 UP000053825 UP000075809 UP000078542 UP000078492 UP000007755 UP000076420 UP000242188 UP000265160 UP000265100 UP000261460 UP000078540 UP000076502 UP000053454 UP000000311 UP000037069 UP000015101 UP000019118 UP000078541 UP000085678 UP000014760 UP000005207 UP000193380 UP000261500 UP000018467 UP000261360 UP000261420 UP000265080 UP000261560 UP000261440 UP000000305 UP000087266 UP000001038 UP000265180 UP000265200 UP000246464
PRIDE
Pfam
PF00335 Tetraspanin
Interpro
SUPFAM
SSF48652
SSF48652
Gene 3D
ProteinModelPortal
H9J0G2
A0A212FA61
A0A2A4JK78
A0A2H1V280
A0A2W1BX49
A0A194PP14
+ More
A0A194QSB2 A0A0L7L883 D6WNZ1 A0A2J7RAC0 A0A1Y1LWZ5 A0A1B0D099 U4TY90 U5EVC2 A0A0T6BC03 A0A2P8XHC9 K7IRI9 A0A087UPS1 A0A1D2NHQ3 A0A2L2YQI9 T1IN95 A0A1J1IQH4 A0A2P6KYT6 A0A2T7NC29 E9IK86 E2BKP2 A0A0C9QX11 V3ZNB1 C3YK57 A0A088ABB0 K1Q8A5 A0A2A3E0X8 V9ICQ6 Q2TCS1 A0A0M9ABC1 A0A0L7QW36 A0A151XG20 A0A195CTI3 A0A151IXH2 F4WYM6 A0A2C9LYG3 A0A210QYA5 A0A3P9BIC1 A0A3P8N992 A0A3B4FXW5 A0A2R5LE09 A0A195BY00 A0A154P8E8 A0A0L8I669 E2AFI6 A0A0L0CFQ3 T1FN76 N6TLA3 A0A195F0D5 A0A146X6Y6 A0A1S3JDE5 R7UTH9 A0A146X895 A0A0C9RC81 A0A034WAS6 I3J346 A0A060X046 A0A3B3VI42 A0A147ACX0 A0A3B1JWB4 A0A3B4X585 A0A3B4T5K5 A0A1A8MND5 A0A3P8SA18 I3JHY6 A0A3B3DL88 A0A060WYB6 A0A3B4CRH3 A0A1A8G6E6 A0A3B4G2L7 A0A3P8RGM8 A0A3P9D4U8 A0A1A8B5H4 A0A3B3DL83 K4GIK4 E9FX18 A0A146MK15 A0A1S3MSP3 A0A1S3RSG3 K4FUU5 H2M7Y3 A0A3P9M7E8 A0A3P9I500 A0A2U9C8B9 A0A0C9Q8W8 A0A3B3DJ63 A0A3B3IC82 V9KQW5 A0A3P9I5F1 A0A3B3I5I1 H2L751
A0A194QSB2 A0A0L7L883 D6WNZ1 A0A2J7RAC0 A0A1Y1LWZ5 A0A1B0D099 U4TY90 U5EVC2 A0A0T6BC03 A0A2P8XHC9 K7IRI9 A0A087UPS1 A0A1D2NHQ3 A0A2L2YQI9 T1IN95 A0A1J1IQH4 A0A2P6KYT6 A0A2T7NC29 E9IK86 E2BKP2 A0A0C9QX11 V3ZNB1 C3YK57 A0A088ABB0 K1Q8A5 A0A2A3E0X8 V9ICQ6 Q2TCS1 A0A0M9ABC1 A0A0L7QW36 A0A151XG20 A0A195CTI3 A0A151IXH2 F4WYM6 A0A2C9LYG3 A0A210QYA5 A0A3P9BIC1 A0A3P8N992 A0A3B4FXW5 A0A2R5LE09 A0A195BY00 A0A154P8E8 A0A0L8I669 E2AFI6 A0A0L0CFQ3 T1FN76 N6TLA3 A0A195F0D5 A0A146X6Y6 A0A1S3JDE5 R7UTH9 A0A146X895 A0A0C9RC81 A0A034WAS6 I3J346 A0A060X046 A0A3B3VI42 A0A147ACX0 A0A3B1JWB4 A0A3B4X585 A0A3B4T5K5 A0A1A8MND5 A0A3P8SA18 I3JHY6 A0A3B3DL88 A0A060WYB6 A0A3B4CRH3 A0A1A8G6E6 A0A3B4G2L7 A0A3P8RGM8 A0A3P9D4U8 A0A1A8B5H4 A0A3B3DL83 K4GIK4 E9FX18 A0A146MK15 A0A1S3MSP3 A0A1S3RSG3 K4FUU5 H2M7Y3 A0A3P9M7E8 A0A3P9I500 A0A2U9C8B9 A0A0C9Q8W8 A0A3B3DJ63 A0A3B3IC82 V9KQW5 A0A3P9I5F1 A0A3B3I5I1 H2L751
Ontologies
GO
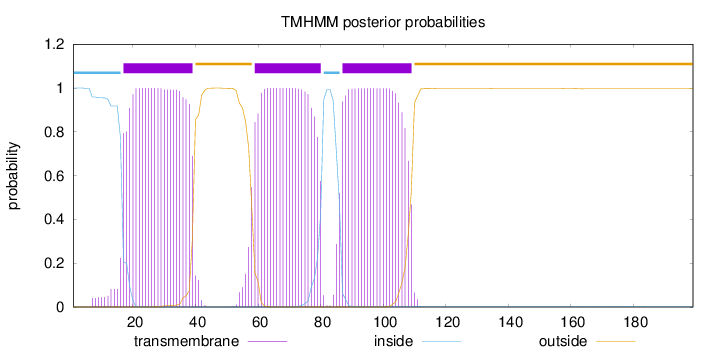
Topology
Subcellular location
Membrane
Length:
199
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.6032500000001
Exp number, first 60 AAs:
25.86044
Total prob of N-in:
0.99962
POSSIBLE N-term signal
sequence
inside
1 - 16
TMhelix
17 - 39
outside
40 - 58
TMhelix
59 - 80
inside
81 - 86
TMhelix
87 - 109
outside
110 - 199
Population Genetic Test Statistics
Pi
175.580505
Theta
197.788411
Tajima's D
-0.379772
CLR
137.243944
CSRT
0.274336283185841
Interpretation
Uncertain
