Pre Gene Modal
BGIBMGA007334
Annotation
putative_NEDD8_precursor_[Danaus_plexippus]
Full name
NEDD8
Alternative Name
Neddylin
Ubiquitin-like protein Nedd8
Ubiquitin-like protein Nedd8
Location in the cell
Cytoplasmic Reliability : 1.281 Mitochondrial Reliability : 1.004 Nuclear Reliability : 1.506
Sequence
CDS
ATGTTGATTAAAGTCAAGACGCTTACTGGTAAAGAAATTGAAATAGATATAGAACCCACCGACAAAGTGGAAAGAATCAAAGAAAGAGTAGAAGAAAAGGAAGGCATTCCGCCGCAGCAACAGCGTCTCATATTTTCAGGAAAACAAATGAATGATGAGAAGACAGCTCAAGATTACAAAGTTCAGGGAGGTTCAGTACTTCATCTTGTTTTGGCTTTAAGGGGAGGCGAATGA
Protein
MLIKVKTLTGKEIEIDIEPTDKVERIKERVEEKEGIPPQQQRLIFSGKQMNDEKTAQDYKVQGGSVLHLVLALRGGE
Summary
Description
Ubiquitin-like protein which plays an important role in cell cycle control, embryogenesis and neurogenesis. Covalent attachment to its substrates requires prior activation by the E1 complex Uba3-Ula1 and linkage to the E2 enzyme Ubc12. Attachment of Nedd8 to cullins activates their associated E3 ubiquitin ligase activity, and thus promotes polyubiquitination and proteasomal degradation of cyclins and other regulatory proteins.
Subunit
Covalently attached to cullins.
Similarity
Belongs to the ubiquitin family.
Keywords
Complete proteome
Developmental protein
Isopeptide bond
Nucleus
Reference proteome
Ubl conjugation pathway
Feature
chain NEDD8
Uniprot
H9JCT7
A0A212ESW7
E0VU21
S4P857
R4V541
A0A2W1BQB3
+ More
A0A2A4JK69 K7J7I0 A0A2A3E9N1 V9IF73 A0A088A321 D6WL56 A0A1W4WES4 J9K6H7 A0A0P4VHF5 A0A069DV91 R4FL16 A0A067RD15 A0A224Y317 A0A158NI45 A0A1B6LAP1 B4MZM8 T1DNZ5 A0A2M4C5A3 A0A182NXB5 A0A023EDB2 A0A2M4ARA0 W5J935 T1DFZ4 Q17NI5 U5ESY7 R4WQJ5 A0A2M3ZI70 B4LSJ8 A0A1J1INB9 A0A182QPK7 A0A1D2N0R3 Q7Q3J6 A0A182IG44 A0A182FWW9 B0W504 B0R023 A0A1L8EAK2 Q6DGU4 A0A1Q3FG27 A0A1S3SDT9 A0A182V579 A0A182L256 B5XE79 A0A3P8TSK8 H2RUL2 B4I5M6 A0A0E9WQ96 B5X5E6 G1FKL5 B4JBY3 A0A1W4VXW6 A0A1B0AUR8 C3KK26 A0A0K8TP95 Q29LT5 B4G7F7 A0A146NPN0 A0A146WH92 H3A9S2 A0A3B0JBK2 G3PMC3 A3KNF0 Q28E73 A0A1S3DU76 A0A0J9TQZ0 X2J935 Q9VJ33 Q4SNR7 H3CR82 A0A0B8RX15 A0A1W7RF99 A0A0K8RYZ5 J3S959 A0A3B1JL70 A0A3B1IJ25 A0A3B3YIE0 A0A087YFD0 A0A3B3UNR2 A0A3P9QEH4 C1BKR4 A0A1S3KDM3 A0A1I8Q3G4 A0A1L8DWV1 A0A1A8NK04 A0A1A8K215 A0A1A8QJR2 A0A1A8DUT5 A0A1A8UZK2 A0A3P9LXQ9 B5X7L9 A0A1A8H196 A0A3B3SAT8 B4PAD3 B3NL79
A0A2A4JK69 K7J7I0 A0A2A3E9N1 V9IF73 A0A088A321 D6WL56 A0A1W4WES4 J9K6H7 A0A0P4VHF5 A0A069DV91 R4FL16 A0A067RD15 A0A224Y317 A0A158NI45 A0A1B6LAP1 B4MZM8 T1DNZ5 A0A2M4C5A3 A0A182NXB5 A0A023EDB2 A0A2M4ARA0 W5J935 T1DFZ4 Q17NI5 U5ESY7 R4WQJ5 A0A2M3ZI70 B4LSJ8 A0A1J1INB9 A0A182QPK7 A0A1D2N0R3 Q7Q3J6 A0A182IG44 A0A182FWW9 B0W504 B0R023 A0A1L8EAK2 Q6DGU4 A0A1Q3FG27 A0A1S3SDT9 A0A182V579 A0A182L256 B5XE79 A0A3P8TSK8 H2RUL2 B4I5M6 A0A0E9WQ96 B5X5E6 G1FKL5 B4JBY3 A0A1W4VXW6 A0A1B0AUR8 C3KK26 A0A0K8TP95 Q29LT5 B4G7F7 A0A146NPN0 A0A146WH92 H3A9S2 A0A3B0JBK2 G3PMC3 A3KNF0 Q28E73 A0A1S3DU76 A0A0J9TQZ0 X2J935 Q9VJ33 Q4SNR7 H3CR82 A0A0B8RX15 A0A1W7RF99 A0A0K8RYZ5 J3S959 A0A3B1JL70 A0A3B1IJ25 A0A3B3YIE0 A0A087YFD0 A0A3B3UNR2 A0A3P9QEH4 C1BKR4 A0A1S3KDM3 A0A1I8Q3G4 A0A1L8DWV1 A0A1A8NK04 A0A1A8K215 A0A1A8QJR2 A0A1A8DUT5 A0A1A8UZK2 A0A3P9LXQ9 B5X7L9 A0A1A8H196 A0A3B3SAT8 B4PAD3 B3NL79
Pubmed
19121390
22118469
20566863
23622113
28756777
20075255
+ More
18362917 19820115 27129103 26334808 24845553 21347285 17994087 24945155 26483478 20920257 23761445 24330624 17510324 23691247 27289101 12364791 14747013 17210077 23594743 20966253 20433749 21551351 25613341 26369729 15632085 9215903 20431018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12231629 15843622 16127432 15496914 25476704 26358130 25727380 23025625 25329095 17554307 29240929 17550304
18362917 19820115 27129103 26334808 24845553 21347285 17994087 24945155 26483478 20920257 23761445 24330624 17510324 23691247 27289101 12364791 14747013 17210077 23594743 20966253 20433749 21551351 25613341 26369729 15632085 9215903 20431018 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12231629 15843622 16127432 15496914 25476704 26358130 25727380 23025625 25329095 17554307 29240929 17550304
EMBL
BABH01027782
AGBW02012723
OWR44544.1
DS235778
EEB16877.1
GAIX01006091
+ More
JAA86469.1 KC741179 AGM33003.1 KZ149933 PZC77259.1 NWSH01001291 PCG71823.1 KZ288322 PBC28184.1 JR040879 AEY59322.1 KQ971343 EFA04065.1 ABLF02031544 GDKW01002705 JAI53890.1 GBGD01003665 JAC85224.1 ACPB03004743 ACPB03004744 GAHY01002279 JAA75231.1 KK852541 KDR21761.1 GFTR01000964 JAW15462.1 ADTU01016271 GEBQ01019195 JAT20782.1 CH963920 EDW77813.1 GAMD01002425 JAA99165.1 GGFJ01011274 MBW60415.1 JXUM01035313 GAPW01006763 GAPW01006762 KQ561054 JAC06836.1 KXJ79855.1 GGFK01009979 MBW43300.1 ADMH02002093 ETN59309.1 GALA01000372 JAA94480.1 CH477199 EAT48231.1 GANO01003008 JAB56863.1 AK417956 BAN21171.1 GGFM01007458 MBW28209.1 CH940649 EDW64820.1 CVRI01000055 CRL01250.1 AXCN02000870 LJIJ01000314 ODM98876.1 AAAB01008964 EAA12768.3 APCN01002503 DS231840 EDS34570.1 FP017218 EF178472 AL772295 ABO92967.1 CAQ14646.1 GFDG01003163 JAV15636.1 BC076245 AAH76245.1 GFDL01008541 JAV26504.1 BT049348 BT149995 ACI69149.1 AGH92599.1 CH480822 EDW55682.1 GBXM01016131 JAH92446.1 BT046265 ACI66066.1 JN216973 AEM37718.1 CH916368 EDW04086.1 JXJN01003720 BT083291 ACQ58998.1 GDAI01001637 JAI15966.1 CH379060 EAL33960.2 CH479180 EDW29290.1 GCES01152937 JAQ33385.1 GCES01058008 JAR28315.1 AFYH01191437 AFYH01191438 OUUW01000004 SPP79415.1 BC133805 AAI33806.1 AAMC01087944 BC122002 CR848418 AAI22003.1 CAJ82981.1 CM002910 KMY90900.1 AE014134 AHN54584.1 BT024445 CAAE01014542 CAF97715.1 GBSH01001779 JAG67247.1 GDAY02001611 JAV49816.1 GBKD01001422 JAG46196.1 JU175219 AFJ50743.1 AYCK01004988 BT075193 ACO09617.1 GFDF01003137 JAV10947.1 HAEG01003423 SBR69204.1 HAEE01006261 SBR26281.1 HAEI01005446 SBR93339.1 HADZ01010556 HAEA01009625 SBQ38105.1 HADY01008820 HAEJ01013058 SBS53515.1 BT047038 ACI66839.1 HAEB01002941 HAEC01008856 SBQ77072.1 CM000158 EDW90341.1 CH954179 EDV54729.1
JAA86469.1 KC741179 AGM33003.1 KZ149933 PZC77259.1 NWSH01001291 PCG71823.1 KZ288322 PBC28184.1 JR040879 AEY59322.1 KQ971343 EFA04065.1 ABLF02031544 GDKW01002705 JAI53890.1 GBGD01003665 JAC85224.1 ACPB03004743 ACPB03004744 GAHY01002279 JAA75231.1 KK852541 KDR21761.1 GFTR01000964 JAW15462.1 ADTU01016271 GEBQ01019195 JAT20782.1 CH963920 EDW77813.1 GAMD01002425 JAA99165.1 GGFJ01011274 MBW60415.1 JXUM01035313 GAPW01006763 GAPW01006762 KQ561054 JAC06836.1 KXJ79855.1 GGFK01009979 MBW43300.1 ADMH02002093 ETN59309.1 GALA01000372 JAA94480.1 CH477199 EAT48231.1 GANO01003008 JAB56863.1 AK417956 BAN21171.1 GGFM01007458 MBW28209.1 CH940649 EDW64820.1 CVRI01000055 CRL01250.1 AXCN02000870 LJIJ01000314 ODM98876.1 AAAB01008964 EAA12768.3 APCN01002503 DS231840 EDS34570.1 FP017218 EF178472 AL772295 ABO92967.1 CAQ14646.1 GFDG01003163 JAV15636.1 BC076245 AAH76245.1 GFDL01008541 JAV26504.1 BT049348 BT149995 ACI69149.1 AGH92599.1 CH480822 EDW55682.1 GBXM01016131 JAH92446.1 BT046265 ACI66066.1 JN216973 AEM37718.1 CH916368 EDW04086.1 JXJN01003720 BT083291 ACQ58998.1 GDAI01001637 JAI15966.1 CH379060 EAL33960.2 CH479180 EDW29290.1 GCES01152937 JAQ33385.1 GCES01058008 JAR28315.1 AFYH01191437 AFYH01191438 OUUW01000004 SPP79415.1 BC133805 AAI33806.1 AAMC01087944 BC122002 CR848418 AAI22003.1 CAJ82981.1 CM002910 KMY90900.1 AE014134 AHN54584.1 BT024445 CAAE01014542 CAF97715.1 GBSH01001779 JAG67247.1 GDAY02001611 JAV49816.1 GBKD01001422 JAG46196.1 JU175219 AFJ50743.1 AYCK01004988 BT075193 ACO09617.1 GFDF01003137 JAV10947.1 HAEG01003423 SBR69204.1 HAEE01006261 SBR26281.1 HAEI01005446 SBR93339.1 HADZ01010556 HAEA01009625 SBQ38105.1 HADY01008820 HAEJ01013058 SBS53515.1 BT047038 ACI66839.1 HAEB01002941 HAEC01008856 SBQ77072.1 CM000158 EDW90341.1 CH954179 EDV54729.1
Proteomes
UP000005204
UP000007151
UP000009046
UP000218220
UP000002358
UP000242457
+ More
UP000005203 UP000007266 UP000192223 UP000007819 UP000015103 UP000027135 UP000005205 UP000007798 UP000075884 UP000069940 UP000249989 UP000000673 UP000008820 UP000008792 UP000183832 UP000075886 UP000094527 UP000007062 UP000075840 UP000069272 UP000002320 UP000000437 UP000087266 UP000075903 UP000075882 UP000265080 UP000005226 UP000001292 UP000001070 UP000192221 UP000092460 UP000001819 UP000008744 UP000008672 UP000268350 UP000007635 UP000008143 UP000079169 UP000000803 UP000007303 UP000018467 UP000261480 UP000028760 UP000261500 UP000242638 UP000085678 UP000095300 UP000265180 UP000265200 UP000261540 UP000002282 UP000008711
UP000005203 UP000007266 UP000192223 UP000007819 UP000015103 UP000027135 UP000005205 UP000007798 UP000075884 UP000069940 UP000249989 UP000000673 UP000008820 UP000008792 UP000183832 UP000075886 UP000094527 UP000007062 UP000075840 UP000069272 UP000002320 UP000000437 UP000087266 UP000075903 UP000075882 UP000265080 UP000005226 UP000001292 UP000001070 UP000192221 UP000092460 UP000001819 UP000008744 UP000008672 UP000268350 UP000007635 UP000008143 UP000079169 UP000000803 UP000007303 UP000018467 UP000261480 UP000028760 UP000261500 UP000242638 UP000085678 UP000095300 UP000265180 UP000265200 UP000261540 UP000002282 UP000008711
Pfam
PF00240 ubiquitin
Interpro
SUPFAM
SSF54236
SSF54236
CDD
ProteinModelPortal
H9JCT7
A0A212ESW7
E0VU21
S4P857
R4V541
A0A2W1BQB3
+ More
A0A2A4JK69 K7J7I0 A0A2A3E9N1 V9IF73 A0A088A321 D6WL56 A0A1W4WES4 J9K6H7 A0A0P4VHF5 A0A069DV91 R4FL16 A0A067RD15 A0A224Y317 A0A158NI45 A0A1B6LAP1 B4MZM8 T1DNZ5 A0A2M4C5A3 A0A182NXB5 A0A023EDB2 A0A2M4ARA0 W5J935 T1DFZ4 Q17NI5 U5ESY7 R4WQJ5 A0A2M3ZI70 B4LSJ8 A0A1J1INB9 A0A182QPK7 A0A1D2N0R3 Q7Q3J6 A0A182IG44 A0A182FWW9 B0W504 B0R023 A0A1L8EAK2 Q6DGU4 A0A1Q3FG27 A0A1S3SDT9 A0A182V579 A0A182L256 B5XE79 A0A3P8TSK8 H2RUL2 B4I5M6 A0A0E9WQ96 B5X5E6 G1FKL5 B4JBY3 A0A1W4VXW6 A0A1B0AUR8 C3KK26 A0A0K8TP95 Q29LT5 B4G7F7 A0A146NPN0 A0A146WH92 H3A9S2 A0A3B0JBK2 G3PMC3 A3KNF0 Q28E73 A0A1S3DU76 A0A0J9TQZ0 X2J935 Q9VJ33 Q4SNR7 H3CR82 A0A0B8RX15 A0A1W7RF99 A0A0K8RYZ5 J3S959 A0A3B1JL70 A0A3B1IJ25 A0A3B3YIE0 A0A087YFD0 A0A3B3UNR2 A0A3P9QEH4 C1BKR4 A0A1S3KDM3 A0A1I8Q3G4 A0A1L8DWV1 A0A1A8NK04 A0A1A8K215 A0A1A8QJR2 A0A1A8DUT5 A0A1A8UZK2 A0A3P9LXQ9 B5X7L9 A0A1A8H196 A0A3B3SAT8 B4PAD3 B3NL79
A0A2A4JK69 K7J7I0 A0A2A3E9N1 V9IF73 A0A088A321 D6WL56 A0A1W4WES4 J9K6H7 A0A0P4VHF5 A0A069DV91 R4FL16 A0A067RD15 A0A224Y317 A0A158NI45 A0A1B6LAP1 B4MZM8 T1DNZ5 A0A2M4C5A3 A0A182NXB5 A0A023EDB2 A0A2M4ARA0 W5J935 T1DFZ4 Q17NI5 U5ESY7 R4WQJ5 A0A2M3ZI70 B4LSJ8 A0A1J1INB9 A0A182QPK7 A0A1D2N0R3 Q7Q3J6 A0A182IG44 A0A182FWW9 B0W504 B0R023 A0A1L8EAK2 Q6DGU4 A0A1Q3FG27 A0A1S3SDT9 A0A182V579 A0A182L256 B5XE79 A0A3P8TSK8 H2RUL2 B4I5M6 A0A0E9WQ96 B5X5E6 G1FKL5 B4JBY3 A0A1W4VXW6 A0A1B0AUR8 C3KK26 A0A0K8TP95 Q29LT5 B4G7F7 A0A146NPN0 A0A146WH92 H3A9S2 A0A3B0JBK2 G3PMC3 A3KNF0 Q28E73 A0A1S3DU76 A0A0J9TQZ0 X2J935 Q9VJ33 Q4SNR7 H3CR82 A0A0B8RX15 A0A1W7RF99 A0A0K8RYZ5 J3S959 A0A3B1JL70 A0A3B1IJ25 A0A3B3YIE0 A0A087YFD0 A0A3B3UNR2 A0A3P9QEH4 C1BKR4 A0A1S3KDM3 A0A1I8Q3G4 A0A1L8DWV1 A0A1A8NK04 A0A1A8K215 A0A1A8QJR2 A0A1A8DUT5 A0A1A8UZK2 A0A3P9LXQ9 B5X7L9 A0A1A8H196 A0A3B3SAT8 B4PAD3 B3NL79
PDB
4HCP
E-value=9.15054e-23,
Score=257
Ontologies
GO
Topology
Subcellular location
Nucleus
Mainly nuclear. With evidence from 1 publications.
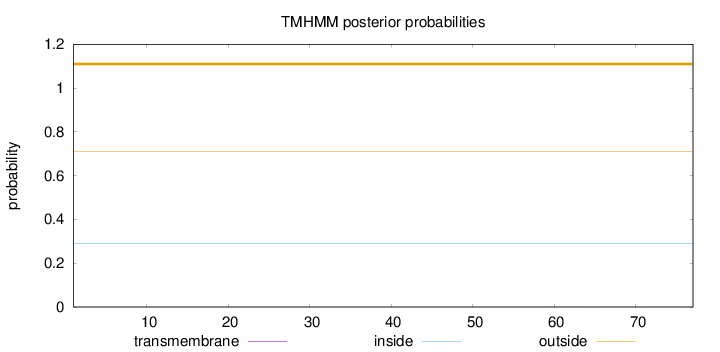
Length:
77
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00245
Exp number, first 60 AAs:
0.0003
Total prob of N-in:
0.29002
outside
1 - 77
Population Genetic Test Statistics
Pi
19.072047
Theta
57.505536
Tajima's D
-1.65361
CLR
880.945327
CSRT
0.0438978051097445
Interpretation
Possibly Positive selection
