Pre Gene Modal
BGIBMGA002990
Annotation
PREDICTED:_sorting_nexin-20_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 2.301
Sequence
CDS
ATGTTGCGATTCAAAATTATTTCGTCACGGACAGTGGATAGTTCTGAGACCGAGAAGAAATTTGTAGCCTACATGGTACAGGTGAATCGAGAAACGGATTTGGCTGATCAAGATCCTGCGAACGTTGAAAGAAGATACACCCATTTTTTGGATCTATATAATGGTTTAAAAAAGGAACAACCGACGCTGCTTAATAATTTATCATTTCCGAGAAAGACTGTAGTAGGCAACTTTGATGCCAATTTAATATCTACTCGTTGTGCAGCATTTGAATCACTGTTGGATTTAATGAGCAATGACTCAAGACTCCGAGATTGTCCGGCAGCAATCACATTCTTCCAGGATGTGGAATTGAGTGAAGCCAAGAGACTAATTAATGAGGGTAAATTTGACCAGGCACTGTCCATACTAGAAACAAGTTTCAAATTATTAAATAAGGTTTATACAGACAGATCAAGGGTGGTGCTTTGTGCTCTTTGTCGGATTGTGGCATGCGCTGGAGCTAGTGATGGTACATTAGCAGGGCCAGTGGAACGGTGGGCACAGCTTGCATTACGAAGATACGAAGCAGTCAGTGATTCTGATTTACTCCTGATATACATACCATTGCTTCACACTTGCATAAACATTTGGGAAACACTTGGTAGAGATAAATCGAAGTTAGTTGAAGAACTAAATGACCTTCGCAAACGAGGAATGAAGGTGGATTCCGTGCCCACATTAATGGAAGCAGTCGACACTTTAGACACCATGTAA
Protein
MLRFKIISSRTVDSSETEKKFVAYMVQVNRETDLADQDPANVERRYTHFLDLYNGLKKEQPTLLNNLSFPRKTVVGNFDANLISTRCAAFESLLDLMSNDSRLRDCPAAITFFQDVELSEAKRLINEGKFDQALSILETSFKLLNKVYTDRSRVVLCALCRIVACAGASDGTLAGPVERWAQLALRRYEAVSDSDLLLIYIPLLHTCINIWETLGRDKSKLVEELNDLRKRGMKVDSVPTLMEAVDTLDTM
Summary
Uniprot
H9J0F3
A0A2H1WJB5
A0A2A4IUM4
A0A2W1BQB4
A0A1E1VZ09
A0A194QN98
+ More
A0A194Q5L3 A0A0L7LW05 A0A212ESV4 A0A067R163 A0A2J7QMA9 A0A2J7QMA6 A0A2J7QMD1 D6WL42 A0A1W4XMK9 A0A2P8YFM8 A0A1B0CAW2 A0A1I8P6T7 A0A1I8P6Z4 A0A0K8VWK8 A0A034WL02 A0A0K8V7X5 A0A336M4Q0 A0A182GGX2 A0A1I8NF49 A0A1I8NF48 A0A023EPM1 A0A1L8DW93 W8BZQ4 W8CA01 Q16S18 W8C5J4 A0A1S4FR48 A0A1B6CRH7 A0A182SRH4 A0A1W4V6N4 U5EV36 A0A1B6D3M3 B4LUU7 A0A0A1XFW1 A0A0A1WLG7 A0A1L8DWA0 A0A1B0FQX7 Q29KW6 B4GSP9 A0A0L0CFZ0 A0A3B0K749 A0A0Q9X808 B4KLD5 A0A1Y1MZU8 A0A1A9V6V8 A0A0Q9XI80 B4NWU2 A0A084VND1 B4Q8K0 A0A0J9QV94 B3N1A5 Q9VQG1 M9PBU4 A0A1A9X608 A0A1B0BWC7 B4I2P8 B3NA55 B4N0H2 A0A069DRI2 B4JQQ2 A0A1J1ICW4 A0A023EYC3 A0A182KBK6 E0VVL8 A0A182MDX9 A0A0V0GCQ4 A0A182PDE0 A0A182QF40 A0A182NEW3 A0A182YFF3 A0A182URL6 A0A1S4H0W8 A0NGB2 A0A2C9GMK5 A0A182WGY5 A0A182X2X0 A0A182I4Y3 A0A182R9Q8 T1IC44 A0A182TIF2 A0A182J311 A0A1B6I1J4 A0A1B6ILK7 A0A2R7VY22 A0A1B6EVN4 A0A1Q3FDE3 A0A1Q3FDI2 A0A224XIZ6 A0A1Q3FDH0 J3JU69 A0A2M4AUP1 A0A2M4AUS6 A0A2M4AU35
A0A194Q5L3 A0A0L7LW05 A0A212ESV4 A0A067R163 A0A2J7QMA9 A0A2J7QMA6 A0A2J7QMD1 D6WL42 A0A1W4XMK9 A0A2P8YFM8 A0A1B0CAW2 A0A1I8P6T7 A0A1I8P6Z4 A0A0K8VWK8 A0A034WL02 A0A0K8V7X5 A0A336M4Q0 A0A182GGX2 A0A1I8NF49 A0A1I8NF48 A0A023EPM1 A0A1L8DW93 W8BZQ4 W8CA01 Q16S18 W8C5J4 A0A1S4FR48 A0A1B6CRH7 A0A182SRH4 A0A1W4V6N4 U5EV36 A0A1B6D3M3 B4LUU7 A0A0A1XFW1 A0A0A1WLG7 A0A1L8DWA0 A0A1B0FQX7 Q29KW6 B4GSP9 A0A0L0CFZ0 A0A3B0K749 A0A0Q9X808 B4KLD5 A0A1Y1MZU8 A0A1A9V6V8 A0A0Q9XI80 B4NWU2 A0A084VND1 B4Q8K0 A0A0J9QV94 B3N1A5 Q9VQG1 M9PBU4 A0A1A9X608 A0A1B0BWC7 B4I2P8 B3NA55 B4N0H2 A0A069DRI2 B4JQQ2 A0A1J1ICW4 A0A023EYC3 A0A182KBK6 E0VVL8 A0A182MDX9 A0A0V0GCQ4 A0A182PDE0 A0A182QF40 A0A182NEW3 A0A182YFF3 A0A182URL6 A0A1S4H0W8 A0NGB2 A0A2C9GMK5 A0A182WGY5 A0A182X2X0 A0A182I4Y3 A0A182R9Q8 T1IC44 A0A182TIF2 A0A182J311 A0A1B6I1J4 A0A1B6ILK7 A0A2R7VY22 A0A1B6EVN4 A0A1Q3FDE3 A0A1Q3FDI2 A0A224XIZ6 A0A1Q3FDH0 J3JU69 A0A2M4AUP1 A0A2M4AUS6 A0A2M4AU35
Pubmed
19121390
28756777
26354079
26227816
22118469
24845553
+ More
18362917 19820115 29403074 25348373 26483478 25315136 24945155 24495485 17510324 17994087 25830018 15632085 26108605 28004739 17550304 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 25474469 20566863 25244985 12364791 22516182
18362917 19820115 29403074 25348373 26483478 25315136 24945155 24495485 17510324 17994087 25830018 15632085 26108605 28004739 17550304 24438588 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 25474469 20566863 25244985 12364791 22516182
EMBL
BABH01010478
ODYU01009031
SOQ53138.1
NWSH01006437
PCG63431.1
KZ149933
+ More
PZC77272.1 GDQN01011081 JAT79973.1 KQ461196 KPJ06450.1 KQ459583 KPI98685.1 JTDY01000025 KOB79351.1 AGBW02012723 OWR44557.1 KK852781 KDR16645.1 NEVH01013211 PNF29730.1 PNF29731.1 PNF29732.1 KQ971343 EFA03513.1 PYGN01000636 PSN43041.1 AJWK01004336 GDHF01032413 GDHF01021340 GDHF01017960 GDHF01009053 GDHF01008540 GDHF01005871 JAI19901.1 JAI30974.1 JAI34354.1 JAI43261.1 JAI43774.1 JAI46443.1 GAKP01004132 JAC54820.1 GDHF01017343 JAI34971.1 UFQT01000439 SSX24331.1 JXUM01062620 JXUM01062621 KQ562208 KXJ76408.1 GAPW01002648 JAC10950.1 GFDF01003386 JAV10698.1 GAMC01001703 JAC04853.1 GAMC01001704 JAC04852.1 CH477689 EAT37250.1 GAMC01001702 JAC04854.1 GEDC01021280 JAS16018.1 GANO01003586 JAB56285.1 GEDC01016999 JAS20299.1 CH940649 EDW64274.2 GBXI01004461 JAD09831.1 GBXI01014952 JAC99339.1 GFDF01003385 JAV10699.1 CCAG010010711 CH379061 EAL33058.3 CH479189 EDW25408.1 JRES01000438 KNC31165.1 OUUW01000006 SPP81849.1 CH933807 KRG03433.1 EDW12816.1 GEZM01016696 JAV91184.1 KRG03432.1 CM000157 EDW87434.1 ATLV01014737 KE524984 KFB39475.1 CM000361 EDX03517.1 CM002910 KMY87729.1 CH902652 EDV33626.2 AE014134 AY061149 AAF51213.2 AAL28697.1 AGB92499.1 JXJN01021707 CH480820 EDW54043.1 CH954177 EDV57518.1 CH963920 EDW77585.2 GBGD01002637 JAC86252.1 CH916372 EDV99232.1 CVRI01000047 CRK97396.1 GBBI01004625 JAC14087.1 DS235812 EEB17424.1 AXCM01011510 GECL01000225 JAP05899.1 AXCN02000839 AAAB01008984 EAU75941.2 APCN01003737 ACPB03011330 GECU01026923 GECU01018131 GECU01017011 GECU01008896 JAS80783.1 JAS89575.1 JAS90695.1 JAS98810.1 GECU01019903 GECU01018359 JAS87803.1 JAS89347.1 KK854163 PTY12397.1 GECZ01027685 JAS42084.1 GFDL01009449 JAV25596.1 GFDL01009425 JAV25620.1 GFTR01003961 JAW12465.1 GFDL01009448 JAV25597.1 BT126782 AEE61744.1 GGFK01011173 MBW44494.1 GGFK01011027 MBW44348.1 GGFK01010969 MBW44290.1
PZC77272.1 GDQN01011081 JAT79973.1 KQ461196 KPJ06450.1 KQ459583 KPI98685.1 JTDY01000025 KOB79351.1 AGBW02012723 OWR44557.1 KK852781 KDR16645.1 NEVH01013211 PNF29730.1 PNF29731.1 PNF29732.1 KQ971343 EFA03513.1 PYGN01000636 PSN43041.1 AJWK01004336 GDHF01032413 GDHF01021340 GDHF01017960 GDHF01009053 GDHF01008540 GDHF01005871 JAI19901.1 JAI30974.1 JAI34354.1 JAI43261.1 JAI43774.1 JAI46443.1 GAKP01004132 JAC54820.1 GDHF01017343 JAI34971.1 UFQT01000439 SSX24331.1 JXUM01062620 JXUM01062621 KQ562208 KXJ76408.1 GAPW01002648 JAC10950.1 GFDF01003386 JAV10698.1 GAMC01001703 JAC04853.1 GAMC01001704 JAC04852.1 CH477689 EAT37250.1 GAMC01001702 JAC04854.1 GEDC01021280 JAS16018.1 GANO01003586 JAB56285.1 GEDC01016999 JAS20299.1 CH940649 EDW64274.2 GBXI01004461 JAD09831.1 GBXI01014952 JAC99339.1 GFDF01003385 JAV10699.1 CCAG010010711 CH379061 EAL33058.3 CH479189 EDW25408.1 JRES01000438 KNC31165.1 OUUW01000006 SPP81849.1 CH933807 KRG03433.1 EDW12816.1 GEZM01016696 JAV91184.1 KRG03432.1 CM000157 EDW87434.1 ATLV01014737 KE524984 KFB39475.1 CM000361 EDX03517.1 CM002910 KMY87729.1 CH902652 EDV33626.2 AE014134 AY061149 AAF51213.2 AAL28697.1 AGB92499.1 JXJN01021707 CH480820 EDW54043.1 CH954177 EDV57518.1 CH963920 EDW77585.2 GBGD01002637 JAC86252.1 CH916372 EDV99232.1 CVRI01000047 CRK97396.1 GBBI01004625 JAC14087.1 DS235812 EEB17424.1 AXCM01011510 GECL01000225 JAP05899.1 AXCN02000839 AAAB01008984 EAU75941.2 APCN01003737 ACPB03011330 GECU01026923 GECU01018131 GECU01017011 GECU01008896 JAS80783.1 JAS89575.1 JAS90695.1 JAS98810.1 GECU01019903 GECU01018359 JAS87803.1 JAS89347.1 KK854163 PTY12397.1 GECZ01027685 JAS42084.1 GFDL01009449 JAV25596.1 GFDL01009425 JAV25620.1 GFTR01003961 JAW12465.1 GFDL01009448 JAV25597.1 BT126782 AEE61744.1 GGFK01011173 MBW44494.1 GGFK01011027 MBW44348.1 GGFK01010969 MBW44290.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000027135 UP000235965 UP000007266 UP000192223 UP000245037 UP000092461 UP000095300 UP000069940 UP000249989 UP000095301 UP000008820 UP000075901 UP000192221 UP000008792 UP000092444 UP000001819 UP000008744 UP000037069 UP000268350 UP000009192 UP000078200 UP000002282 UP000030765 UP000000304 UP000007801 UP000000803 UP000092443 UP000092460 UP000001292 UP000008711 UP000007798 UP000001070 UP000183832 UP000075881 UP000009046 UP000075883 UP000075885 UP000075886 UP000075884 UP000076408 UP000075903 UP000007062 UP000075840 UP000075920 UP000076407 UP000075900 UP000015103 UP000075902 UP000075880
UP000027135 UP000235965 UP000007266 UP000192223 UP000245037 UP000092461 UP000095300 UP000069940 UP000249989 UP000095301 UP000008820 UP000075901 UP000192221 UP000008792 UP000092444 UP000001819 UP000008744 UP000037069 UP000268350 UP000009192 UP000078200 UP000002282 UP000030765 UP000000304 UP000007801 UP000000803 UP000092443 UP000092460 UP000001292 UP000008711 UP000007798 UP000001070 UP000183832 UP000075881 UP000009046 UP000075883 UP000075885 UP000075886 UP000075884 UP000076408 UP000075903 UP000007062 UP000075840 UP000075920 UP000076407 UP000075900 UP000015103 UP000075902 UP000075880
Pfam
PF00787 PX
Interpro
Gene 3D
ProteinModelPortal
H9J0F3
A0A2H1WJB5
A0A2A4IUM4
A0A2W1BQB4
A0A1E1VZ09
A0A194QN98
+ More
A0A194Q5L3 A0A0L7LW05 A0A212ESV4 A0A067R163 A0A2J7QMA9 A0A2J7QMA6 A0A2J7QMD1 D6WL42 A0A1W4XMK9 A0A2P8YFM8 A0A1B0CAW2 A0A1I8P6T7 A0A1I8P6Z4 A0A0K8VWK8 A0A034WL02 A0A0K8V7X5 A0A336M4Q0 A0A182GGX2 A0A1I8NF49 A0A1I8NF48 A0A023EPM1 A0A1L8DW93 W8BZQ4 W8CA01 Q16S18 W8C5J4 A0A1S4FR48 A0A1B6CRH7 A0A182SRH4 A0A1W4V6N4 U5EV36 A0A1B6D3M3 B4LUU7 A0A0A1XFW1 A0A0A1WLG7 A0A1L8DWA0 A0A1B0FQX7 Q29KW6 B4GSP9 A0A0L0CFZ0 A0A3B0K749 A0A0Q9X808 B4KLD5 A0A1Y1MZU8 A0A1A9V6V8 A0A0Q9XI80 B4NWU2 A0A084VND1 B4Q8K0 A0A0J9QV94 B3N1A5 Q9VQG1 M9PBU4 A0A1A9X608 A0A1B0BWC7 B4I2P8 B3NA55 B4N0H2 A0A069DRI2 B4JQQ2 A0A1J1ICW4 A0A023EYC3 A0A182KBK6 E0VVL8 A0A182MDX9 A0A0V0GCQ4 A0A182PDE0 A0A182QF40 A0A182NEW3 A0A182YFF3 A0A182URL6 A0A1S4H0W8 A0NGB2 A0A2C9GMK5 A0A182WGY5 A0A182X2X0 A0A182I4Y3 A0A182R9Q8 T1IC44 A0A182TIF2 A0A182J311 A0A1B6I1J4 A0A1B6ILK7 A0A2R7VY22 A0A1B6EVN4 A0A1Q3FDE3 A0A1Q3FDI2 A0A224XIZ6 A0A1Q3FDH0 J3JU69 A0A2M4AUP1 A0A2M4AUS6 A0A2M4AU35
A0A194Q5L3 A0A0L7LW05 A0A212ESV4 A0A067R163 A0A2J7QMA9 A0A2J7QMA6 A0A2J7QMD1 D6WL42 A0A1W4XMK9 A0A2P8YFM8 A0A1B0CAW2 A0A1I8P6T7 A0A1I8P6Z4 A0A0K8VWK8 A0A034WL02 A0A0K8V7X5 A0A336M4Q0 A0A182GGX2 A0A1I8NF49 A0A1I8NF48 A0A023EPM1 A0A1L8DW93 W8BZQ4 W8CA01 Q16S18 W8C5J4 A0A1S4FR48 A0A1B6CRH7 A0A182SRH4 A0A1W4V6N4 U5EV36 A0A1B6D3M3 B4LUU7 A0A0A1XFW1 A0A0A1WLG7 A0A1L8DWA0 A0A1B0FQX7 Q29KW6 B4GSP9 A0A0L0CFZ0 A0A3B0K749 A0A0Q9X808 B4KLD5 A0A1Y1MZU8 A0A1A9V6V8 A0A0Q9XI80 B4NWU2 A0A084VND1 B4Q8K0 A0A0J9QV94 B3N1A5 Q9VQG1 M9PBU4 A0A1A9X608 A0A1B0BWC7 B4I2P8 B3NA55 B4N0H2 A0A069DRI2 B4JQQ2 A0A1J1ICW4 A0A023EYC3 A0A182KBK6 E0VVL8 A0A182MDX9 A0A0V0GCQ4 A0A182PDE0 A0A182QF40 A0A182NEW3 A0A182YFF3 A0A182URL6 A0A1S4H0W8 A0NGB2 A0A2C9GMK5 A0A182WGY5 A0A182X2X0 A0A182I4Y3 A0A182R9Q8 T1IC44 A0A182TIF2 A0A182J311 A0A1B6I1J4 A0A1B6ILK7 A0A2R7VY22 A0A1B6EVN4 A0A1Q3FDE3 A0A1Q3FDI2 A0A224XIZ6 A0A1Q3FDH0 J3JU69 A0A2M4AUP1 A0A2M4AUS6 A0A2M4AU35
PDB
1XTE
E-value=0.000532474,
Score=100
Ontologies
GO
PANTHER
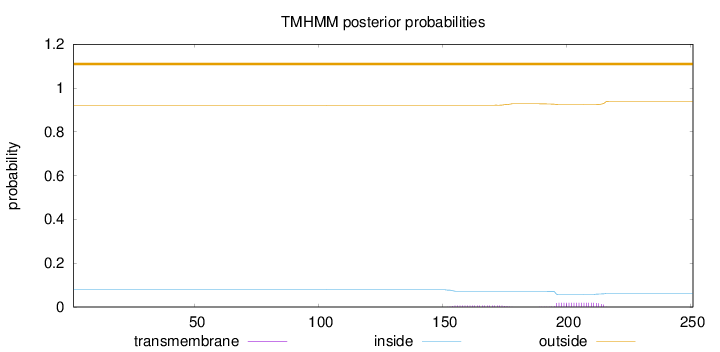
Topology
Length:
251
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.52651
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07837
outside
1 - 251
Population Genetic Test Statistics
Pi
280.578298
Theta
219.132183
Tajima's D
0.959454
CLR
0.122484
CSRT
0.656417179141043
Interpretation
Uncertain
