Gene
KWMTBOMO02101
Pre Gene Modal
BGIBMGA002988
Annotation
PREDICTED:_large_neutral_amino_acids_transporter_small_subunit_2_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 4.966
Sequence
CDS
ATGGGAACGATTATTGGATCCGGAATATTCATATCCCCTGCTGGAGTTTACTTGTACACAGGGTCAGTGGCTGCCTCGTTAATAATATGGCTTGCGAGCGGACTTCTGTCCACGCTGGGAGCTTTGTGCTACGCCGAGCTGGGTACATCGATCACAAGATCAGGAGGAGACTATGCGTACATATATACAGCCTTCGGACCGCTGCCGGCGTTTTTGAGGCTCTGGATTGCACTGCTCATTATCAGGCCTACCACCCAGGCGATAGTGGCGCTCACCTTTGGAAATTACGTTGTCAAACCCTTCTTTCCGGAGTGTGATCCACCGGAGAACGCTGTCAGACTCCTTGCTGCAGTATGCTTGTGTGTACTGACTGCGATAAACTGTATCAGCGTGAGATGGACCATGCGCATCCAAGATGTGTTTACAACGTCAAAGCTACTGGCTCTAGTCGTCATCATTATTTCGGGACTCTACTATATTTGCATAGGTAACACACAAAATTTCGAGAACGCATTTGCTGGAGAATATAGTGCTGGTAATATTGCGCTAGCCTTTTATTCGGGCCTCTTCGCTTTTGGAGGTTGGAATTATCTCAACTTTGTAACCGAAGAACTCCAGGATCCTTATAAGAATCTTCCTCGCGCGATATGGATCGCTTTGCCTCTGGTTACGATTATCTACGTCATGGCAAACTTGGCCTACTTTGCGGTCGTCACTAAAATGGAAATGATGGCAAATCCAGCCGTTGCTGCGATATTCGGTGATCGCCTCTTCGGAAACTGGAGCTGGGTGATTCCAGTGTTCGTGGCGTTGTCCACTTTTGGAGGCGTCAACGGTGTCTTATTCACGTCAGCTCGTCTATTCGCCACCGGCGCCCAGGAGGGACACATGCCGGCGTTCTTTTCACTGTTCCACGTCGATAAACAAACTCCTATACCCTCTCTTATATTTACGTGTTTCTTCTCGTTACTCATGTTGACAACTAGCAACGTTTTCGCCCTTATAAACTACTACTCGCAGATACTTTGGCTTTCCGTCGGCGCATCTGTCGTTGGAATGTTGTGGCTTCGACGAACGAAACCTGACCTGCCTCGGCCAATCAAAGTCAACATAGTTATACCGTACATATTTCTCATCGCTATTGCGTGTTTAGTTATAATTCCGGCGATTGTACAGCCCAAGGATACTGCAATAGGTATTGTTATTTTACTCTCTGGCATCCCTGTTTACTATTTGTGTGTAAAATGGCAAAGGAAACCAGAAGCGTATCATTCGTTCTCCGGTGGCATTTTGAGGTTCCTACAAAAAACATGTTCCTGTATTTATCTTGATTCCACAGAGAAATTGTCTGAGAGTTAG
Protein
MGTIIGSGIFISPAGVYLYTGSVAASLIIWLASGLLSTLGALCYAELGTSITRSGGDYAYIYTAFGPLPAFLRLWIALLIIRPTTQAIVALTFGNYVVKPFFPECDPPENAVRLLAAVCLCVLTAINCISVRWTMRIQDVFTTSKLLALVVIIISGLYYICIGNTQNFENAFAGEYSAGNIALAFYSGLFAFGGWNYLNFVTEELQDPYKNLPRAIWIALPLVTIIYVMANLAYFAVVTKMEMMANPAVAAIFGDRLFGNWSWVIPVFVALSTFGGVNGVLFTSARLFATGAQEGHMPAFFSLFHVDKQTPIPSLIFTCFFSLLMLTTSNVFALINYYSQILWLSVGASVVGMLWLRRTKPDLPRPIKVNIVIPYIFLIAIACLVIIPAIVQPKDTAIGIVILLSGIPVYYLCVKWQRKPEAYHSFSGGILRFLQKTCSCIYLDSTEKLSES
Summary
Uniprot
A0A2W1BUF8
A0A194QLP9
A0A212ESU3
A0A0M3QTS8
B4N798
A0A0L0C8U3
+ More
A0A1B6KXX4 A0A0K8U6B8 A0A034W7R9 B4LUH5 B4KHZ0 D6WNJ1 A0A1B6D0D7 B4P1X7 B4JBK6 B3N3Y1 B4GQJ3 Q29NJ9 A0A3B0K607 A0A1W4UYQ0 Q9VKC2 W8AMB0 A0A0A1X787 B4IE55 A0A1B0BI19 A0A1A9XUE9 A0A1B6ILP1 A0A1B6FVA4 B3MVI6 A0A1A9WZS2 A0A1I8PVX4 A0A182LZK6 A0A1Y9GLJ8 B4Q3P2 A0A1A9UGV2 A0A2M4AAC5 A0A2M4AAP9 A0A1A9ZR29 A0A182QSE9 A0A1I8N6J1 U5ETJ4 A0A1B0G797 A0A2M3YYU9 A0A2M3YYT6 Q17E71 A0A182JIN8 A0A084W7T9 S4P8K7 A0A1Q3FLB6 A0A1J1J311 A0A1Q3FL71 B0W124 U4TZP9 A0A067QPT5 A0A1Y1N544 R4FMC0 A0A224XFU9 A0A146LGY9 A0A0A9Y5T6 N6U6I6 E0VVE6 A0A023ERZ6 R4WDM1 A0A2P8YUZ1 T1J266 A0A131YP25 A0A224Z1S0 R7V9S2 A0A293LYV5 A0A1B0CTD0 A0A087U4Z8 A0A0L8H4R7 A0A2R5LNY6 A0A1Z5L7Y8 A0A2M4AAW9 Q7QKR6 V4A3A4 A0A154PMR5 A0A2M4ABG0 E2BKM3 A0A182VPP9 A0A182T518 A0A0B6ZJE7 A0A182REC1 A0A2M4ABR2 B5DYS6 A0A232ERK8 A0A1L8DEG0 B4GYT3 K7IXD9 A0A210PSU8 A0A182NMK2 A0A182FP70 A0A1W7RA21 T1EFM3 A0A2L2YCD9 A0A0R3NFJ2 A0A182K8S7 A0A182PQA6 K1Q3X1
A0A1B6KXX4 A0A0K8U6B8 A0A034W7R9 B4LUH5 B4KHZ0 D6WNJ1 A0A1B6D0D7 B4P1X7 B4JBK6 B3N3Y1 B4GQJ3 Q29NJ9 A0A3B0K607 A0A1W4UYQ0 Q9VKC2 W8AMB0 A0A0A1X787 B4IE55 A0A1B0BI19 A0A1A9XUE9 A0A1B6ILP1 A0A1B6FVA4 B3MVI6 A0A1A9WZS2 A0A1I8PVX4 A0A182LZK6 A0A1Y9GLJ8 B4Q3P2 A0A1A9UGV2 A0A2M4AAC5 A0A2M4AAP9 A0A1A9ZR29 A0A182QSE9 A0A1I8N6J1 U5ETJ4 A0A1B0G797 A0A2M3YYU9 A0A2M3YYT6 Q17E71 A0A182JIN8 A0A084W7T9 S4P8K7 A0A1Q3FLB6 A0A1J1J311 A0A1Q3FL71 B0W124 U4TZP9 A0A067QPT5 A0A1Y1N544 R4FMC0 A0A224XFU9 A0A146LGY9 A0A0A9Y5T6 N6U6I6 E0VVE6 A0A023ERZ6 R4WDM1 A0A2P8YUZ1 T1J266 A0A131YP25 A0A224Z1S0 R7V9S2 A0A293LYV5 A0A1B0CTD0 A0A087U4Z8 A0A0L8H4R7 A0A2R5LNY6 A0A1Z5L7Y8 A0A2M4AAW9 Q7QKR6 V4A3A4 A0A154PMR5 A0A2M4ABG0 E2BKM3 A0A182VPP9 A0A182T518 A0A0B6ZJE7 A0A182REC1 A0A2M4ABR2 B5DYS6 A0A232ERK8 A0A1L8DEG0 B4GYT3 K7IXD9 A0A210PSU8 A0A182NMK2 A0A182FP70 A0A1W7RA21 T1EFM3 A0A2L2YCD9 A0A0R3NFJ2 A0A182K8S7 A0A182PQA6 K1Q3X1
Pubmed
28756777
26354079
22118469
17994087
26108605
25348373
+ More
18057021 18362917 19820115 17550304 15632085 23185243 10731132 12537568 12537572 12537573 12537574 12530223 16110336 17569856 17569867 26109357 26109356 24495485 25830018 22936249 25315136 17510324 24438588 23622113 23537049 24845553 28004739 26823975 25401762 20566863 24945155 23691247 29403074 26830274 28797301 23254933 28528879 9087549 12364791 14747013 17210077 20798317 28648823 20075255 28812685 26561354 22992520
18057021 18362917 19820115 17550304 15632085 23185243 10731132 12537568 12537572 12537573 12537574 12530223 16110336 17569856 17569867 26109357 26109356 24495485 25830018 22936249 25315136 17510324 24438588 23622113 23537049 24845553 28004739 26823975 25401762 20566863 24945155 23691247 29403074 26830274 28797301 23254933 28528879 9087549 12364791 14747013 17210077 20798317 28648823 20075255 28812685 26561354 22992520
EMBL
KZ149933
PZC77274.1
KQ461196
KPJ06452.1
AGBW02012723
OWR44558.1
+ More
CP012523 ALC39409.1 CH964182 EDW80239.1 JRES01000841 KNC27814.1 GEBQ01023664 JAT16313.1 GDHF01031919 GDHF01030159 GDHF01030120 GDHF01024280 GDHF01020508 JAI20395.1 JAI22155.1 JAI22194.1 JAI28034.1 JAI31806.1 GAKP01008595 GAKP01008594 GAKP01008593 JAC50357.1 CH940649 EDW64161.1 CH933807 EDW11272.1 KRG02625.1 KQ971343 EFA03221.1 GEDC01018109 JAS19189.1 CM000157 EDW88148.1 KRJ97606.1 CH916368 EDW04029.1 CH954177 EDV58833.1 CH479187 EDW39865.1 CH379059 EAL34220.1 KRT03755.1 OUUW01000006 SPP81439.1 AE014134 AF273478 AY051907 AAF53154.1 AAK58692.1 AAK93331.1 AAN10800.2 AFH03670.1 AHN54377.1 AHN54378.1 GAMC01020979 GAMC01020976 GAMC01020973 GAMC01020970 GAMC01020967 JAB85588.1 GBXI01007754 JAD06538.1 CH480831 EDW45882.1 JXJN01014746 GECU01019872 JAS87834.1 GECZ01015680 JAS54089.1 CH902624 EDV33251.1 KPU74366.1 KPU74367.1 AXCM01000109 APCN01005972 CM000361 CM002910 EDX04767.1 KMY89865.1 GGFK01004371 MBW37692.1 GGFK01004524 MBW37845.1 AXCN02001594 GANO01002754 JAB57117.1 CCAG010022298 GGFM01000699 MBW21450.1 GGFM01000678 MBW21429.1 CH477285 EAT44748.1 ATLV01021297 KE525316 KFB46283.1 GAIX01005886 JAA86674.1 GFDL01006666 JAV28379.1 CVRI01000067 CRL06751.1 GFDL01006685 JAV28360.1 DS231819 EDS42841.1 KB631904 ERL87094.1 KK853096 KDR11465.1 GEZM01013433 JAV92558.1 ACPB03012598 ACPB03012599 GAHY01001769 JAA75741.1 GFTR01006512 JAW09914.1 GDHC01011205 JAQ07424.1 GBHO01040858 GBHO01015177 GBHO01000716 GBHO01000113 GBHO01000108 JAG02746.1 JAG28427.1 JAG42888.1 JAG43491.1 JAG43496.1 APGK01040748 APGK01040749 APGK01040750 KB740984 ENN76266.1 DS235811 EEB17352.1 GAPW01001601 JAC11997.1 AK417813 BAN21028.1 PYGN01000345 PSN48047.1 JH431796 GEDV01008239 JAP80318.1 GFPF01011969 MAA23115.1 AMQN01018059 KB293833 ELU15339.1 GFWV01008410 MAA33139.1 AJWK01027431 KK118198 KFM72437.1 KQ419199 KOF84286.1 GGLE01007127 MBY11253.1 GFJQ02003590 JAW03380.1 GGFK01004612 MBW37933.1 AAAB01008555 EAA03429.3 KB202823 ESO87801.1 KQ434991 KZC13175.1 GGFK01004627 MBW37948.1 GL448819 EFN83721.1 HACG01021783 CEK68648.1 GGFK01004869 MBW38190.1 CM000070 EDY67636.2 NNAY01002581 OXU20999.1 GFDF01009235 JAV04849.1 CH479198 EDW27951.1 AAZX01000012 NEDP02005521 OWF39558.1 GFAH01000390 JAV47999.1 AMQM01005099 KB096785 ESO01412.1 IAAA01013998 IAAA01013999 LAA05030.1 KRS99769.1 KRS99770.1 JH818799 EKC28548.1
CP012523 ALC39409.1 CH964182 EDW80239.1 JRES01000841 KNC27814.1 GEBQ01023664 JAT16313.1 GDHF01031919 GDHF01030159 GDHF01030120 GDHF01024280 GDHF01020508 JAI20395.1 JAI22155.1 JAI22194.1 JAI28034.1 JAI31806.1 GAKP01008595 GAKP01008594 GAKP01008593 JAC50357.1 CH940649 EDW64161.1 CH933807 EDW11272.1 KRG02625.1 KQ971343 EFA03221.1 GEDC01018109 JAS19189.1 CM000157 EDW88148.1 KRJ97606.1 CH916368 EDW04029.1 CH954177 EDV58833.1 CH479187 EDW39865.1 CH379059 EAL34220.1 KRT03755.1 OUUW01000006 SPP81439.1 AE014134 AF273478 AY051907 AAF53154.1 AAK58692.1 AAK93331.1 AAN10800.2 AFH03670.1 AHN54377.1 AHN54378.1 GAMC01020979 GAMC01020976 GAMC01020973 GAMC01020970 GAMC01020967 JAB85588.1 GBXI01007754 JAD06538.1 CH480831 EDW45882.1 JXJN01014746 GECU01019872 JAS87834.1 GECZ01015680 JAS54089.1 CH902624 EDV33251.1 KPU74366.1 KPU74367.1 AXCM01000109 APCN01005972 CM000361 CM002910 EDX04767.1 KMY89865.1 GGFK01004371 MBW37692.1 GGFK01004524 MBW37845.1 AXCN02001594 GANO01002754 JAB57117.1 CCAG010022298 GGFM01000699 MBW21450.1 GGFM01000678 MBW21429.1 CH477285 EAT44748.1 ATLV01021297 KE525316 KFB46283.1 GAIX01005886 JAA86674.1 GFDL01006666 JAV28379.1 CVRI01000067 CRL06751.1 GFDL01006685 JAV28360.1 DS231819 EDS42841.1 KB631904 ERL87094.1 KK853096 KDR11465.1 GEZM01013433 JAV92558.1 ACPB03012598 ACPB03012599 GAHY01001769 JAA75741.1 GFTR01006512 JAW09914.1 GDHC01011205 JAQ07424.1 GBHO01040858 GBHO01015177 GBHO01000716 GBHO01000113 GBHO01000108 JAG02746.1 JAG28427.1 JAG42888.1 JAG43491.1 JAG43496.1 APGK01040748 APGK01040749 APGK01040750 KB740984 ENN76266.1 DS235811 EEB17352.1 GAPW01001601 JAC11997.1 AK417813 BAN21028.1 PYGN01000345 PSN48047.1 JH431796 GEDV01008239 JAP80318.1 GFPF01011969 MAA23115.1 AMQN01018059 KB293833 ELU15339.1 GFWV01008410 MAA33139.1 AJWK01027431 KK118198 KFM72437.1 KQ419199 KOF84286.1 GGLE01007127 MBY11253.1 GFJQ02003590 JAW03380.1 GGFK01004612 MBW37933.1 AAAB01008555 EAA03429.3 KB202823 ESO87801.1 KQ434991 KZC13175.1 GGFK01004627 MBW37948.1 GL448819 EFN83721.1 HACG01021783 CEK68648.1 GGFK01004869 MBW38190.1 CM000070 EDY67636.2 NNAY01002581 OXU20999.1 GFDF01009235 JAV04849.1 CH479198 EDW27951.1 AAZX01000012 NEDP02005521 OWF39558.1 GFAH01000390 JAV47999.1 AMQM01005099 KB096785 ESO01412.1 IAAA01013998 IAAA01013999 LAA05030.1 KRS99769.1 KRS99770.1 JH818799 EKC28548.1
Proteomes
UP000053240
UP000007151
UP000092553
UP000007798
UP000037069
UP000008792
+ More
UP000009192 UP000007266 UP000002282 UP000001070 UP000008711 UP000008744 UP000001819 UP000268350 UP000192221 UP000000803 UP000001292 UP000092460 UP000092443 UP000007801 UP000091820 UP000095300 UP000075883 UP000075840 UP000000304 UP000078200 UP000092445 UP000075886 UP000095301 UP000092444 UP000008820 UP000075880 UP000030765 UP000183832 UP000002320 UP000030742 UP000027135 UP000015103 UP000019118 UP000009046 UP000245037 UP000014760 UP000092461 UP000054359 UP000053454 UP000007062 UP000030746 UP000076502 UP000008237 UP000075920 UP000075901 UP000075900 UP000215335 UP000002358 UP000242188 UP000075884 UP000069272 UP000015101 UP000075881 UP000075885 UP000005408
UP000009192 UP000007266 UP000002282 UP000001070 UP000008711 UP000008744 UP000001819 UP000268350 UP000192221 UP000000803 UP000001292 UP000092460 UP000092443 UP000007801 UP000091820 UP000095300 UP000075883 UP000075840 UP000000304 UP000078200 UP000092445 UP000075886 UP000095301 UP000092444 UP000008820 UP000075880 UP000030765 UP000183832 UP000002320 UP000030742 UP000027135 UP000015103 UP000019118 UP000009046 UP000245037 UP000014760 UP000092461 UP000054359 UP000053454 UP000007062 UP000030746 UP000076502 UP000008237 UP000075920 UP000075901 UP000075900 UP000215335 UP000002358 UP000242188 UP000075884 UP000069272 UP000015101 UP000075881 UP000075885 UP000005408
PRIDE
Pfam
PF13520 AA_permease_2
Interpro
IPR002293
AA/rel_permease1
ProteinModelPortal
A0A2W1BUF8
A0A194QLP9
A0A212ESU3
A0A0M3QTS8
B4N798
A0A0L0C8U3
+ More
A0A1B6KXX4 A0A0K8U6B8 A0A034W7R9 B4LUH5 B4KHZ0 D6WNJ1 A0A1B6D0D7 B4P1X7 B4JBK6 B3N3Y1 B4GQJ3 Q29NJ9 A0A3B0K607 A0A1W4UYQ0 Q9VKC2 W8AMB0 A0A0A1X787 B4IE55 A0A1B0BI19 A0A1A9XUE9 A0A1B6ILP1 A0A1B6FVA4 B3MVI6 A0A1A9WZS2 A0A1I8PVX4 A0A182LZK6 A0A1Y9GLJ8 B4Q3P2 A0A1A9UGV2 A0A2M4AAC5 A0A2M4AAP9 A0A1A9ZR29 A0A182QSE9 A0A1I8N6J1 U5ETJ4 A0A1B0G797 A0A2M3YYU9 A0A2M3YYT6 Q17E71 A0A182JIN8 A0A084W7T9 S4P8K7 A0A1Q3FLB6 A0A1J1J311 A0A1Q3FL71 B0W124 U4TZP9 A0A067QPT5 A0A1Y1N544 R4FMC0 A0A224XFU9 A0A146LGY9 A0A0A9Y5T6 N6U6I6 E0VVE6 A0A023ERZ6 R4WDM1 A0A2P8YUZ1 T1J266 A0A131YP25 A0A224Z1S0 R7V9S2 A0A293LYV5 A0A1B0CTD0 A0A087U4Z8 A0A0L8H4R7 A0A2R5LNY6 A0A1Z5L7Y8 A0A2M4AAW9 Q7QKR6 V4A3A4 A0A154PMR5 A0A2M4ABG0 E2BKM3 A0A182VPP9 A0A182T518 A0A0B6ZJE7 A0A182REC1 A0A2M4ABR2 B5DYS6 A0A232ERK8 A0A1L8DEG0 B4GYT3 K7IXD9 A0A210PSU8 A0A182NMK2 A0A182FP70 A0A1W7RA21 T1EFM3 A0A2L2YCD9 A0A0R3NFJ2 A0A182K8S7 A0A182PQA6 K1Q3X1
A0A1B6KXX4 A0A0K8U6B8 A0A034W7R9 B4LUH5 B4KHZ0 D6WNJ1 A0A1B6D0D7 B4P1X7 B4JBK6 B3N3Y1 B4GQJ3 Q29NJ9 A0A3B0K607 A0A1W4UYQ0 Q9VKC2 W8AMB0 A0A0A1X787 B4IE55 A0A1B0BI19 A0A1A9XUE9 A0A1B6ILP1 A0A1B6FVA4 B3MVI6 A0A1A9WZS2 A0A1I8PVX4 A0A182LZK6 A0A1Y9GLJ8 B4Q3P2 A0A1A9UGV2 A0A2M4AAC5 A0A2M4AAP9 A0A1A9ZR29 A0A182QSE9 A0A1I8N6J1 U5ETJ4 A0A1B0G797 A0A2M3YYU9 A0A2M3YYT6 Q17E71 A0A182JIN8 A0A084W7T9 S4P8K7 A0A1Q3FLB6 A0A1J1J311 A0A1Q3FL71 B0W124 U4TZP9 A0A067QPT5 A0A1Y1N544 R4FMC0 A0A224XFU9 A0A146LGY9 A0A0A9Y5T6 N6U6I6 E0VVE6 A0A023ERZ6 R4WDM1 A0A2P8YUZ1 T1J266 A0A131YP25 A0A224Z1S0 R7V9S2 A0A293LYV5 A0A1B0CTD0 A0A087U4Z8 A0A0L8H4R7 A0A2R5LNY6 A0A1Z5L7Y8 A0A2M4AAW9 Q7QKR6 V4A3A4 A0A154PMR5 A0A2M4ABG0 E2BKM3 A0A182VPP9 A0A182T518 A0A0B6ZJE7 A0A182REC1 A0A2M4ABR2 B5DYS6 A0A232ERK8 A0A1L8DEG0 B4GYT3 K7IXD9 A0A210PSU8 A0A182NMK2 A0A182FP70 A0A1W7RA21 T1EFM3 A0A2L2YCD9 A0A0R3NFJ2 A0A182K8S7 A0A182PQA6 K1Q3X1
PDB
6IRT
E-value=9.03182e-112,
Score=1032
Ontologies
GO
Topology
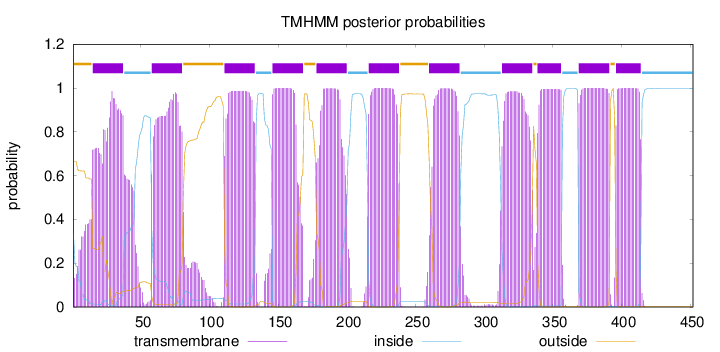
Length:
452
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
244.29844
Exp number, first 60 AAs:
30.76898
Total prob of N-in:
0.33568
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 37
inside
38 - 57
TMhelix
58 - 80
outside
81 - 110
TMhelix
111 - 133
inside
134 - 145
TMhelix
146 - 168
outside
169 - 177
TMhelix
178 - 200
inside
201 - 215
TMhelix
216 - 238
outside
239 - 259
TMhelix
260 - 282
inside
283 - 312
TMhelix
313 - 335
outside
336 - 338
TMhelix
339 - 356
inside
357 - 368
TMhelix
369 - 391
outside
392 - 395
TMhelix
396 - 414
inside
415 - 452
Population Genetic Test Statistics
Pi
237.744961
Theta
20.843617
Tajima's D
0.394305
CLR
0.597447
CSRT
0.485425728713564
Interpretation
Uncertain
