Pre Gene Modal
BGIBMGA003178
Annotation
PREDICTED:_large_neutral_amino_acids_transporter_small_subunit_1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.989
Sequence
CDS
ATGGCTAAAGTTGCCGACGTAGATGGATTAGCTCCCAAGGCTATCGAAAATGAAGTTGAAAGCAGTGATGGTGTCGAAAAAGGATCTGGTGGAGGTGTGCGATTGAAAAAAGAGTTGAGTCTAATGAACGGTGTGGCAATAATTGTGGGTGTAATAGTCGGATCAGGAATATTCGTTTCTCCAAGTTTGGCGCTAAAACATGCAGGTTCTAAAGGAATGGCTCTCATCGTGTGGGTTTTATCAGGATTCTTGTCCATGATAGGAGCGCTCTGTTATGCAGAGCTAGGTACGATGATCCCGAAATCTGGTGGGGATTACGCGTATATCGGTGAAGCGTTTGGTTCTCTACCGGCTTTTCTCTATCTGTGGGTTGCGCTCTTCATCCTTGTACCGACGGGAAACGCAATCACAGCCCTGACCTTCGCTGAAAACATCCTGAAGCCGTTATGGCCAGTTTGCAATCCTCCGGTCGTAGCAGTCAACCTTATTGCAGCCAGCATTACCTGTTTCCTGACAATAATTAACTGTTATAACGTGAAATGGGTGACGCGAGTTCAAGACTCGTTTACCGCTGCGAAAGTACTGGCGCTACTCGTGACATTCTTCGCGAGCTTGGTCTACTTATTCTCCGGACACACGGAAAACCTGCAGTACATGATGGAGAAAACAACAACGGATCCAGGGGAGATCGCTATAGCATTTTATACCGGCCTATTTTCGTATTCTGGCTGGAATTACCTGAACTTTGTTACTGAAGAACTGAAGGATCCCTACAAGAATCTTCCACGAGCTATCTGCATATCTATGCCGGTCGTGACCTTGGTCTACACGCTAACCAACGTCGCCTATTTCGCTGTATTGTCCAGTGACGAAATCCTTTCGTCCTCCGCCGTCGCTGTTACGTTCAGCGAGAAAATTCTCAAAATGATGTCTTGGATAATGCCGCTCTTCGTGGCTCTGTGTACATTTGGATCCCTCAACGGCGCCATATACACTTCCTCGCGACTTTTCTTCGTCGGTGCCAGGAACGGACATTTACCTTTGGCGATTTCACTCATAGACATCAAGAGGTTGACCCCGGTGCCTTCACTCATATTTATGTGCCTGGTCACCCTGCTGCTACTGCTGTCGAACGACATCGAAGCTCTGATGGTGTACGTGACTGCTGTCGAAGCGTTGTTCACCCTCTGCTCGGTGACCGGCTTGCTGTGGATGCGCTACACGCGACCACGCCTCCAGCGACCGATCCGCGTCAGTCTCGTCCTGCCCGCTATCTTCCTGATAACCTGCACATTTATTGTCATATGCTCGTGCTTTAAATACCCGAAGCACGTCGGCATCGGGGTCGCCTTCATAGCGCTGGGCGTCCCCATATACATGATATTCATCAAATGGCAGAATAAACCCAATTGGATATTAACAGCGTGCAATAGCTTCAATTTGGCGTGTTCCAAGCTGTTTTTGTGTTTACCCGAGGACTGCAAAGAATTATGA
Protein
MAKVADVDGLAPKAIENEVESSDGVEKGSGGGVRLKKELSLMNGVAIIVGVIVGSGIFVSPSLALKHAGSKGMALIVWVLSGFLSMIGALCYAELGTMIPKSGGDYAYIGEAFGSLPAFLYLWVALFILVPTGNAITALTFAENILKPLWPVCNPPVVAVNLIAASITCFLTIINCYNVKWVTRVQDSFTAAKVLALLVTFFASLVYLFSGHTENLQYMMEKTTTDPGEIAIAFYTGLFSYSGWNYLNFVTEELKDPYKNLPRAICISMPVVTLVYTLTNVAYFAVLSSDEILSSSAVAVTFSEKILKMMSWIMPLFVALCTFGSLNGAIYTSSRLFFVGARNGHLPLAISLIDIKRLTPVPSLIFMCLVTLLLLLSNDIEALMVYVTAVEALFTLCSVTGLLWMRYTRPRLQRPIRVSLVLPAIFLITCTFIVICSCFKYPKHVGIGVAFIALGVPIYMIFIKWQNKPNWILTACNSFNLACSKLFLCLPEDCKEL
Summary
Uniprot
H9J0Z1
A0A2A4J4H0
A0A2W1BQJ7
A0A194Q070
A0A194QSD7
A0A2H1X2C0
+ More
A0A212FJV9 A0A1Y1KA98 N6U9P6 A0A232ERK8 E2BKM3 K7IXD9 A0A0L7QM92 A0A1I8NCV7 A0A1I8QED9 A0A0L0C122 A0A0K8U881 B4J2W6 A0A2A3E9V0 A0A088A296 A0A1B0GAQ3 A0A1A9YTC4 A0A1B0A6E0 A0A026WC57 W8AKL5 A0A1W4XEE1 A0A1Y0AWM1 U5EHX8 A0A1A9WTC3 D6WN13 E2A792 E0VEN0 A0A151I831 A0A336MVI9 A0A1B6D737 A0A158NJZ0 B3M5Q7 A0A154PMR5 A0A084WUS2 B4LFL6 A0A1A9VU07 A0A336M249 B4KXM6 A0A2J7QMB7 A0A182N371 B4PII9 B3NHK4 A0A1W4U8W7 A0A1J1J6S5 B4HHV5 Q9Y1A7 A0A3B0JTL0 B4GUH1 A0A195BHJ3 B5DQR8 B4QKH9 A0A3L8DNF1 A0A195DZ38 B4NM02 A0A0J9RWK9 F4WIK2 A0A151XEG5 A0A182QT17 A0A182IM50 A0A182W2C6 A0A182URT0 A0A2C9H3X7 A0A182LQ77 A0A182PAX5 A0A182HY00 A0A182LWL5 W5JDW3 A0A182UL39 A0A023F2Z4 Q16H52 A0A182R571 A0A182H8R0 A0A1L8DE69 A0A182KC11 A0A182GAG3 A0A1Q3G0T4 A0A1Q3G108 A0A1Q3G0H3 A0A1Q3G0L4 A0A1Q3G0Z7 A0A1Q3G0W5 A0A2M4A2S5 A0A1Q3G0J0 A0A1Q3G0K3 Q7Q2X6 A0A2M3Z3G9 A0A2M3ZGQ6 A0A182YMG2 T1DLD8 A0A023ETB0 A0A182FBS2 E9JCD9 A0A146L8N6 A0A0A9YVI6 A0A310SJC6
A0A212FJV9 A0A1Y1KA98 N6U9P6 A0A232ERK8 E2BKM3 K7IXD9 A0A0L7QM92 A0A1I8NCV7 A0A1I8QED9 A0A0L0C122 A0A0K8U881 B4J2W6 A0A2A3E9V0 A0A088A296 A0A1B0GAQ3 A0A1A9YTC4 A0A1B0A6E0 A0A026WC57 W8AKL5 A0A1W4XEE1 A0A1Y0AWM1 U5EHX8 A0A1A9WTC3 D6WN13 E2A792 E0VEN0 A0A151I831 A0A336MVI9 A0A1B6D737 A0A158NJZ0 B3M5Q7 A0A154PMR5 A0A084WUS2 B4LFL6 A0A1A9VU07 A0A336M249 B4KXM6 A0A2J7QMB7 A0A182N371 B4PII9 B3NHK4 A0A1W4U8W7 A0A1J1J6S5 B4HHV5 Q9Y1A7 A0A3B0JTL0 B4GUH1 A0A195BHJ3 B5DQR8 B4QKH9 A0A3L8DNF1 A0A195DZ38 B4NM02 A0A0J9RWK9 F4WIK2 A0A151XEG5 A0A182QT17 A0A182IM50 A0A182W2C6 A0A182URT0 A0A2C9H3X7 A0A182LQ77 A0A182PAX5 A0A182HY00 A0A182LWL5 W5JDW3 A0A182UL39 A0A023F2Z4 Q16H52 A0A182R571 A0A182H8R0 A0A1L8DE69 A0A182KC11 A0A182GAG3 A0A1Q3G0T4 A0A1Q3G108 A0A1Q3G0H3 A0A1Q3G0L4 A0A1Q3G0Z7 A0A1Q3G0W5 A0A2M4A2S5 A0A1Q3G0J0 A0A1Q3G0K3 Q7Q2X6 A0A2M3Z3G9 A0A2M3ZGQ6 A0A182YMG2 T1DLD8 A0A023ETB0 A0A182FBS2 E9JCD9 A0A146L8N6 A0A0A9YVI6 A0A310SJC6
Pubmed
19121390
28756777
26354079
22118469
28004739
23537049
+ More
28648823 20798317 20075255 25315136 26108605 17994087 24508170 24495485 28341416 18362917 19820115 20566863 21347285 24438588 17550304 10727855 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 30249741 22936249 21719571 12364791 20966253 20920257 23761445 25474469 17510324 26483478 25244985 24945155 21282665 26823975 25401762
28648823 20798317 20075255 25315136 26108605 17994087 24508170 24495485 28341416 18362917 19820115 20566863 21347285 24438588 17550304 10727855 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 30249741 22936249 21719571 12364791 20966253 20920257 23761445 25474469 17510324 26483478 25244985 24945155 21282665 26823975 25401762
EMBL
BABH01010482
BABH01010483
NWSH01003137
PCG66891.1
KZ149933
PZC77278.1
+ More
KQ459583 KPI98693.1 KQ461196 KPJ06456.1 ODYU01012917 SOQ59459.1 AGBW02008222 OWR53990.1 GEZM01092432 GEZM01092431 JAV56555.1 APGK01037602 APGK01037603 KB740948 KB632288 ENN77371.1 ERL91483.1 NNAY01002581 OXU20999.1 GL448819 EFN83721.1 AAZX01000012 KQ414902 KOC59646.1 JRES01001055 KNC25957.1 GDHF01031203 GDHF01029447 GDHF01022579 GDHF01001097 JAI21111.1 JAI22867.1 JAI29735.1 JAI51217.1 CH916366 EDV97136.1 KZ288311 PBC28498.1 CCAG010015501 KK107282 EZA53558.1 GAMC01020188 GAMC01020184 JAB86367.1 KY921794 ART29383.1 GANO01002844 JAB57027.1 KQ971343 EFA03252.1 GL437314 EFN70702.1 DS235093 EEB11836.1 KQ978390 KYM94322.1 UFQT01002849 SSX34140.1 GEDC01015923 JAS21375.1 ADTU01018478 ADTU01018479 ADTU01018480 CH902618 EDV40691.1 KQ434991 KZC13175.1 ATLV01027105 KE525423 KFB53966.1 CH940647 EDW70334.1 UFQT01000416 SSX24120.1 CH933809 EDW19733.1 NEVH01013211 PNF29741.1 CM000159 EDW94546.1 CH954178 EDV51799.1 CVRI01000070 CRL07182.1 CH480815 EDW41520.1 AF139834 AE014296 AY069533 AAD39459.1 AAF49688.1 AAL39678.1 AAN11781.3 AAN11782.1 OUUW01000002 SPP76031.1 CH479191 EDW26254.1 KQ976465 KYM84275.1 CH379069 EDY73384.1 KRT07543.1 CM000363 EDX10490.1 QOIP01000006 RLU21964.1 KQ980031 KYN18155.1 CH964278 EDW85370.1 CM002912 KMY99643.1 KMY99644.1 GL888176 EGI65938.1 KQ982254 KYQ58648.1 AXCN02001123 AAAB01008966 APCN01004447 APCN01004448 AXCM01009262 ADMH02001789 ETN61085.1 GBBI01003124 JAC15588.1 CH478204 EAT33566.1 JXUM01029435 KQ560853 KXJ80622.1 GFDF01009409 JAV04675.1 JXUM01050863 KQ561665 KXJ77871.1 GFDL01001634 JAV33411.1 GFDL01001561 JAV33484.1 GFDL01001741 JAV33304.1 GFDL01001718 JAV33327.1 GFDL01001577 JAV33468.1 GFDL01001600 JAV33445.1 GGFK01001720 MBW35041.1 GFDL01001724 JAV33321.1 GFDL01001749 JAV33296.1 EAA13031.4 GGFM01002294 MBW23045.1 GGFM01006930 MBW27681.1 GAMD01000501 JAB01090.1 GAPW01001033 JAC12565.1 GL771786 EFZ09530.1 GDHC01013845 JAQ04784.1 GBHO01006557 JAG37047.1 KQ761976 OAD56413.1
KQ459583 KPI98693.1 KQ461196 KPJ06456.1 ODYU01012917 SOQ59459.1 AGBW02008222 OWR53990.1 GEZM01092432 GEZM01092431 JAV56555.1 APGK01037602 APGK01037603 KB740948 KB632288 ENN77371.1 ERL91483.1 NNAY01002581 OXU20999.1 GL448819 EFN83721.1 AAZX01000012 KQ414902 KOC59646.1 JRES01001055 KNC25957.1 GDHF01031203 GDHF01029447 GDHF01022579 GDHF01001097 JAI21111.1 JAI22867.1 JAI29735.1 JAI51217.1 CH916366 EDV97136.1 KZ288311 PBC28498.1 CCAG010015501 KK107282 EZA53558.1 GAMC01020188 GAMC01020184 JAB86367.1 KY921794 ART29383.1 GANO01002844 JAB57027.1 KQ971343 EFA03252.1 GL437314 EFN70702.1 DS235093 EEB11836.1 KQ978390 KYM94322.1 UFQT01002849 SSX34140.1 GEDC01015923 JAS21375.1 ADTU01018478 ADTU01018479 ADTU01018480 CH902618 EDV40691.1 KQ434991 KZC13175.1 ATLV01027105 KE525423 KFB53966.1 CH940647 EDW70334.1 UFQT01000416 SSX24120.1 CH933809 EDW19733.1 NEVH01013211 PNF29741.1 CM000159 EDW94546.1 CH954178 EDV51799.1 CVRI01000070 CRL07182.1 CH480815 EDW41520.1 AF139834 AE014296 AY069533 AAD39459.1 AAF49688.1 AAL39678.1 AAN11781.3 AAN11782.1 OUUW01000002 SPP76031.1 CH479191 EDW26254.1 KQ976465 KYM84275.1 CH379069 EDY73384.1 KRT07543.1 CM000363 EDX10490.1 QOIP01000006 RLU21964.1 KQ980031 KYN18155.1 CH964278 EDW85370.1 CM002912 KMY99643.1 KMY99644.1 GL888176 EGI65938.1 KQ982254 KYQ58648.1 AXCN02001123 AAAB01008966 APCN01004447 APCN01004448 AXCM01009262 ADMH02001789 ETN61085.1 GBBI01003124 JAC15588.1 CH478204 EAT33566.1 JXUM01029435 KQ560853 KXJ80622.1 GFDF01009409 JAV04675.1 JXUM01050863 KQ561665 KXJ77871.1 GFDL01001634 JAV33411.1 GFDL01001561 JAV33484.1 GFDL01001741 JAV33304.1 GFDL01001718 JAV33327.1 GFDL01001577 JAV33468.1 GFDL01001600 JAV33445.1 GGFK01001720 MBW35041.1 GFDL01001724 JAV33321.1 GFDL01001749 JAV33296.1 EAA13031.4 GGFM01002294 MBW23045.1 GGFM01006930 MBW27681.1 GAMD01000501 JAB01090.1 GAPW01001033 JAC12565.1 GL771786 EFZ09530.1 GDHC01013845 JAQ04784.1 GBHO01006557 JAG37047.1 KQ761976 OAD56413.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000019118
+ More
UP000030742 UP000215335 UP000008237 UP000002358 UP000053825 UP000095301 UP000095300 UP000037069 UP000001070 UP000242457 UP000005203 UP000092444 UP000092443 UP000092445 UP000053097 UP000192223 UP000091820 UP000007266 UP000000311 UP000009046 UP000078542 UP000005205 UP000007801 UP000076502 UP000030765 UP000008792 UP000078200 UP000009192 UP000235965 UP000075884 UP000002282 UP000008711 UP000192221 UP000183832 UP000001292 UP000000803 UP000268350 UP000008744 UP000078540 UP000001819 UP000000304 UP000279307 UP000078492 UP000007798 UP000007755 UP000075809 UP000075886 UP000075880 UP000075920 UP000075903 UP000075882 UP000075885 UP000075840 UP000075883 UP000000673 UP000075902 UP000008820 UP000075900 UP000069940 UP000249989 UP000075881 UP000007062 UP000076408 UP000069272
UP000030742 UP000215335 UP000008237 UP000002358 UP000053825 UP000095301 UP000095300 UP000037069 UP000001070 UP000242457 UP000005203 UP000092444 UP000092443 UP000092445 UP000053097 UP000192223 UP000091820 UP000007266 UP000000311 UP000009046 UP000078542 UP000005205 UP000007801 UP000076502 UP000030765 UP000008792 UP000078200 UP000009192 UP000235965 UP000075884 UP000002282 UP000008711 UP000192221 UP000183832 UP000001292 UP000000803 UP000268350 UP000008744 UP000078540 UP000001819 UP000000304 UP000279307 UP000078492 UP000007798 UP000007755 UP000075809 UP000075886 UP000075880 UP000075920 UP000075903 UP000075882 UP000075885 UP000075840 UP000075883 UP000000673 UP000075902 UP000008820 UP000075900 UP000069940 UP000249989 UP000075881 UP000007062 UP000076408 UP000069272
ProteinModelPortal
H9J0Z1
A0A2A4J4H0
A0A2W1BQJ7
A0A194Q070
A0A194QSD7
A0A2H1X2C0
+ More
A0A212FJV9 A0A1Y1KA98 N6U9P6 A0A232ERK8 E2BKM3 K7IXD9 A0A0L7QM92 A0A1I8NCV7 A0A1I8QED9 A0A0L0C122 A0A0K8U881 B4J2W6 A0A2A3E9V0 A0A088A296 A0A1B0GAQ3 A0A1A9YTC4 A0A1B0A6E0 A0A026WC57 W8AKL5 A0A1W4XEE1 A0A1Y0AWM1 U5EHX8 A0A1A9WTC3 D6WN13 E2A792 E0VEN0 A0A151I831 A0A336MVI9 A0A1B6D737 A0A158NJZ0 B3M5Q7 A0A154PMR5 A0A084WUS2 B4LFL6 A0A1A9VU07 A0A336M249 B4KXM6 A0A2J7QMB7 A0A182N371 B4PII9 B3NHK4 A0A1W4U8W7 A0A1J1J6S5 B4HHV5 Q9Y1A7 A0A3B0JTL0 B4GUH1 A0A195BHJ3 B5DQR8 B4QKH9 A0A3L8DNF1 A0A195DZ38 B4NM02 A0A0J9RWK9 F4WIK2 A0A151XEG5 A0A182QT17 A0A182IM50 A0A182W2C6 A0A182URT0 A0A2C9H3X7 A0A182LQ77 A0A182PAX5 A0A182HY00 A0A182LWL5 W5JDW3 A0A182UL39 A0A023F2Z4 Q16H52 A0A182R571 A0A182H8R0 A0A1L8DE69 A0A182KC11 A0A182GAG3 A0A1Q3G0T4 A0A1Q3G108 A0A1Q3G0H3 A0A1Q3G0L4 A0A1Q3G0Z7 A0A1Q3G0W5 A0A2M4A2S5 A0A1Q3G0J0 A0A1Q3G0K3 Q7Q2X6 A0A2M3Z3G9 A0A2M3ZGQ6 A0A182YMG2 T1DLD8 A0A023ETB0 A0A182FBS2 E9JCD9 A0A146L8N6 A0A0A9YVI6 A0A310SJC6
A0A212FJV9 A0A1Y1KA98 N6U9P6 A0A232ERK8 E2BKM3 K7IXD9 A0A0L7QM92 A0A1I8NCV7 A0A1I8QED9 A0A0L0C122 A0A0K8U881 B4J2W6 A0A2A3E9V0 A0A088A296 A0A1B0GAQ3 A0A1A9YTC4 A0A1B0A6E0 A0A026WC57 W8AKL5 A0A1W4XEE1 A0A1Y0AWM1 U5EHX8 A0A1A9WTC3 D6WN13 E2A792 E0VEN0 A0A151I831 A0A336MVI9 A0A1B6D737 A0A158NJZ0 B3M5Q7 A0A154PMR5 A0A084WUS2 B4LFL6 A0A1A9VU07 A0A336M249 B4KXM6 A0A2J7QMB7 A0A182N371 B4PII9 B3NHK4 A0A1W4U8W7 A0A1J1J6S5 B4HHV5 Q9Y1A7 A0A3B0JTL0 B4GUH1 A0A195BHJ3 B5DQR8 B4QKH9 A0A3L8DNF1 A0A195DZ38 B4NM02 A0A0J9RWK9 F4WIK2 A0A151XEG5 A0A182QT17 A0A182IM50 A0A182W2C6 A0A182URT0 A0A2C9H3X7 A0A182LQ77 A0A182PAX5 A0A182HY00 A0A182LWL5 W5JDW3 A0A182UL39 A0A023F2Z4 Q16H52 A0A182R571 A0A182H8R0 A0A1L8DE69 A0A182KC11 A0A182GAG3 A0A1Q3G0T4 A0A1Q3G108 A0A1Q3G0H3 A0A1Q3G0L4 A0A1Q3G0Z7 A0A1Q3G0W5 A0A2M4A2S5 A0A1Q3G0J0 A0A1Q3G0K3 Q7Q2X6 A0A2M3Z3G9 A0A2M3ZGQ6 A0A182YMG2 T1DLD8 A0A023ETB0 A0A182FBS2 E9JCD9 A0A146L8N6 A0A0A9YVI6 A0A310SJC6
PDB
6IRT
E-value=8.01291e-119,
Score=1094
Ontologies
GO
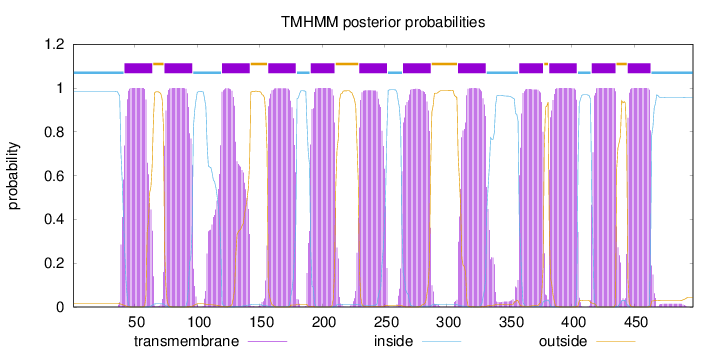
Topology
Length:
497
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
259.06081
Exp number, first 60 AAs:
19.44696
Total prob of N-in:
0.98456
POSSIBLE N-term signal
sequence
inside
1 - 41
TMhelix
42 - 64
outside
65 - 73
TMhelix
74 - 96
inside
97 - 119
TMhelix
120 - 142
outside
143 - 156
TMhelix
157 - 179
inside
180 - 190
TMhelix
191 - 210
outside
211 - 229
TMhelix
230 - 252
inside
253 - 264
TMhelix
265 - 287
outside
288 - 308
TMhelix
309 - 331
inside
332 - 357
TMhelix
358 - 377
outside
378 - 381
TMhelix
382 - 404
inside
405 - 415
TMhelix
416 - 435
outside
436 - 444
TMhelix
445 - 463
inside
464 - 497
Population Genetic Test Statistics
Pi
240.623768
Theta
189.54012
Tajima's D
0.218143
CLR
0.38982
CSRT
0.437078146092695
Interpretation
Uncertain
