Gene
KWMTBOMO02095
Pre Gene Modal
BGIBMGA003180
Annotation
hypothetical_protein_KGM_09574_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 3.907
Sequence
CDS
ATGTTGACAAAGTGCGCCGTCGTAGTCCTCGCGCTGACGCTCGTAGCTGCCGCCGTCTCGGCTAACTCCGAGCTAGACCTCAACAACACTATACCGGCGGAGCGATACTTCATTGACAAGATATTCAACAAGTACGGAGACAAAGGCATCATTACGTTTGAGGGATTCGAACACTTGTTAGCAAGTTTAGGCCTTGGCGGACGCGTGTTCAACTCACCTCATGATCTGGCCCTGCATCGCATCAATGGAACCTTTAGGCCACTTCACGACACCTTGCACAGACACCGACGCTCTCCGCCAGAAATTAGCTCTACAACCATACTACAGAAATCATGCCTTTCACCGAGAGAAATATTGGAAGTGTACGGAATGGAAAACGAGCCCGGGGTGATTACCATAAGACCGAAAACATTCTTGGAGATGTGTCCTGCACTTGTTTATCAATTGGATCAACGTTCGTGTTACAAAACTGAAGTATCTCCACCTAAATTAGATAAAATTTGGACCTGGATTTACGCCACCTTAGCAATTTTAGTGATTAGTGCCACGGGACTCCTGGGTGTAGCAATTGTCCCGCTACTAAAGTCAAAAATATTTAGCCACATATTGCATTTTCTGGTAGCTGTAGCCGTCGGAACACTTTGTGGCGACGCACTGCTACATCTTCTACCACATGCGTTGAAATCTCATGGTCCACCATCAGAACCGCAAGACGAAACAGAAGTGGTGCTTAAATGTAGCGTCACTTTTATCACAATTCTTTTCTTTTACACTGTTGAAGCGATCATGCAAACTGTCAATGGAGGTCATGCTCACTCACACGACCAAAATAAAGGTATTGAAGAAATCAAAACAGACTCGACCAAACAAATTGAGCCTATAGAATTAGGTGCTATGATGCCTGGGTCGCCTCCGCCGCCAATTGAACGACCAATGTCGTCCACTGCTTTAATGGTAATCGTTGGTGATGGATTACATAATCTGACTGATGGTTTGGCAATAGGAGCTGCGTTCAGTGGCGATCCGGTCACAGGATTTGCTACAGCTTTAGCAGTTTTCTGCCACGAGCTGCCACATGAACTGGGAGACTTCGCGGTACTACTCCGATCTGGTATGAGCATCAAGCGTGCTCTGTATTACAACTTACTATCTTCTGTGCTCAGCTTCATGGGAATGGCAGGCGGCATCTGGCTCGCTGAAGACCACGAATCGGCGTCACAGTGGATATACGCGGCGACCGCTGGAACTTTCTTTTACATCGCACTCGCCGATTTAGTTCCAGAAATAAATGAAAATAATAAAGGCAAAAGTGTTAACTTATTACTTGCGGTAATGGGTTTTCTAGCAGGCGGTGTGATCATGTTGCTCATTGCGTTACACGAAGACTCTATACAATATCTGTTTAGAAATGAAGAGAAATAA
Protein
MLTKCAVVVLALTLVAAAVSANSELDLNNTIPAERYFIDKIFNKYGDKGIITFEGFEHLLASLGLGGRVFNSPHDLALHRINGTFRPLHDTLHRHRRSPPEISSTTILQKSCLSPREILEVYGMENEPGVITIRPKTFLEMCPALVYQLDQRSCYKTEVSPPKLDKIWTWIYATLAILVISATGLLGVAIVPLLKSKIFSHILHFLVAVAVGTLCGDALLHLLPHALKSHGPPSEPQDETEVVLKCSVTFITILFFYTVEAIMQTVNGGHAHSHDQNKGIEEIKTDSTKQIEPIELGAMMPGSPPPPIERPMSSTALMVIVGDGLHNLTDGLAIGAAFSGDPVTGFATALAVFCHELPHELGDFAVLLRSGMSIKRALYYNLLSSVLSFMGMAGGIWLAEDHESASQWIYAATAGTFFYIALADLVPEINENNKGKSVNLLLAVMGFLAGGVIMLLIALHEDSIQYLFRNEEK
Summary
Uniprot
A0A2H1X0G2
A0A2W1BQC4
A0A212EN13
A0A194Q129
A0A194QLQ2
H9J0Z3
+ More
A0A2A4IUM0 A0A0L7LV96 A0A067QYY1 A0A2J7RQ13 A0A224XCT1 A0A336M5G6 A0A2J7RQ10 A0A1L8DEX8 B4KXM5 A0A3B0JQU3 B4PIJ0 B4LFL5 A0A1L8DDM2 B4J2W7 B4QKI0 B3NHK5 A4IJ72 Q2M185 B4GUH0 B4HHV6 A0A3Q0J130 A0A1S4ED83 Q16ZM6 A0A1B0CJR3 Q16H51 A0A034VIU8 A0A1S4FIR7 A0A1B6I5Q9 A0A1I8PXS8 A0A1B6KMR7 B0XDI9 E0VTQ8 A0A0Q9WJC4 A0A1B6F1J0 A0A1Q3G554 A0A182GYE9 A0A1Q3G520 A0A1Q3G562 A0A1Q3G533 A0A0M5J5T8 A0A1Q3G539 B3M5Q6 A0A0R1DY55 A0A1Q3G531 W8BTE1 A0A336MYD4 A0A0A9ZAA5 A0A2M4BFL0 A0A2M4BFI7 Q7QLH2 A0A1B0GNR5 A0A1I8MYG1 Q7Q2X7 A0A146L5P5 T1P9P1 A0A0A9ZGZ4 A0A1S4H6I8 A0A2M4CPQ2 A0A1W4UMK0 A0A1A9WQ12 A0A182K1N3 A0A2M4CPU4 A0A2M4CPN0 A0A182FBS3 W5JE42 A0A182JFC6 A0A182LRW7 A0A182QJL7 A0A182HXZ9 B4NM01 A0A182N370 A0A1A9V5R9 A0A182W2C5 A0A2R7VWA6 A0A084WUS0 A0A182PAX6 A0A3B4EJM1 A0A2G8LJ35 A0A2D0PZX4 A0A3B1KKP0 A0A2R8Q6G1 F8W5D4 A0A0P7XSD3 A0A2D3E2E7 A0A3B1JPH6 A0A089ZW72 A0A2M3ZEV5 A0A1W5AQG3 A0A3P8Z3L9 A0A3N0YM80
A0A2A4IUM0 A0A0L7LV96 A0A067QYY1 A0A2J7RQ13 A0A224XCT1 A0A336M5G6 A0A2J7RQ10 A0A1L8DEX8 B4KXM5 A0A3B0JQU3 B4PIJ0 B4LFL5 A0A1L8DDM2 B4J2W7 B4QKI0 B3NHK5 A4IJ72 Q2M185 B4GUH0 B4HHV6 A0A3Q0J130 A0A1S4ED83 Q16ZM6 A0A1B0CJR3 Q16H51 A0A034VIU8 A0A1S4FIR7 A0A1B6I5Q9 A0A1I8PXS8 A0A1B6KMR7 B0XDI9 E0VTQ8 A0A0Q9WJC4 A0A1B6F1J0 A0A1Q3G554 A0A182GYE9 A0A1Q3G520 A0A1Q3G562 A0A1Q3G533 A0A0M5J5T8 A0A1Q3G539 B3M5Q6 A0A0R1DY55 A0A1Q3G531 W8BTE1 A0A336MYD4 A0A0A9ZAA5 A0A2M4BFL0 A0A2M4BFI7 Q7QLH2 A0A1B0GNR5 A0A1I8MYG1 Q7Q2X7 A0A146L5P5 T1P9P1 A0A0A9ZGZ4 A0A1S4H6I8 A0A2M4CPQ2 A0A1W4UMK0 A0A1A9WQ12 A0A182K1N3 A0A2M4CPU4 A0A2M4CPN0 A0A182FBS3 W5JE42 A0A182JFC6 A0A182LRW7 A0A182QJL7 A0A182HXZ9 B4NM01 A0A182N370 A0A1A9V5R9 A0A182W2C5 A0A2R7VWA6 A0A084WUS0 A0A182PAX6 A0A3B4EJM1 A0A2G8LJ35 A0A2D0PZX4 A0A3B1KKP0 A0A2R8Q6G1 F8W5D4 A0A0P7XSD3 A0A2D3E2E7 A0A3B1JPH6 A0A089ZW72 A0A2M3ZEV5 A0A1W5AQG3 A0A3P8Z3L9 A0A3N0YM80
Pubmed
28756777
22118469
26354079
19121390
26227816
24845553
+ More
17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17510324 25348373 20566863 26483478 24495485 25401762 9087549 12364791 14747013 17210077 25315136 26823975 20920257 23761445 24438588 29023486 25329095 23594743 25069045
17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 17510324 25348373 20566863 26483478 24495485 25401762 9087549 12364791 14747013 17210077 25315136 26823975 20920257 23761445 24438588 29023486 25329095 23594743 25069045
EMBL
ODYU01012037
SOQ58124.1
KZ149933
PZC77279.1
AGBW02013744
OWR42884.1
+ More
KQ459583 KPI98694.1 KQ461196 KPJ06457.1 BABH01010486 NWSH01006457 PCG63421.1 JTDY01000025 KOB79355.1 KK852820 KDR15587.1 NEVH01001351 PNF42922.1 GFTR01006180 JAW10246.1 UFQT01000416 SSX24119.1 PNF42925.1 GFDF01009200 JAV04884.1 CH933809 EDW19732.1 OUUW01000002 SPP76029.1 CM000159 EDW94547.1 CH940647 EDW70333.1 GFDF01009546 JAV04538.1 CH916366 EDV97137.1 CM000363 CM002912 EDX10491.1 KMY99645.1 CH954178 EDV51800.1 AE014296 BT030430 AAF49687.2 ABO52850.1 ABW08542.1 CH379069 EAL30690.2 CH479191 EDW26253.1 CH480815 EDW41521.1 CH477488 EAT40100.1 AJWK01014885 CH478204 EAT33567.1 GAKP01016896 GAKP01016895 JAC42057.1 GECU01025438 JAS82268.1 GEBQ01027228 JAT12749.1 DS232762 EDS45497.1 DS235771 EEB16764.1 KRF84861.1 GECZ01029884 GECZ01026048 GECZ01025808 GECZ01017396 GECZ01014368 GECZ01012569 GECZ01011268 GECZ01008724 GECZ01007339 JAS39885.1 JAS43721.1 JAS43961.1 JAS52373.1 JAS55401.1 JAS57200.1 JAS58501.1 JAS61045.1 JAS62430.1 GFDL01000105 JAV34940.1 JXUM01021127 KQ560592 KXJ81745.1 GFDL01000145 JAV34900.1 GFDL01000139 JAV34906.1 GFDL01000138 JAV34907.1 CP012525 ALC43818.1 GFDL01000125 JAV34920.1 CH902618 EDV40690.1 KRK01909.1 GFDL01000141 JAV34904.1 GAMC01010074 JAB96481.1 UFQT01002849 SSX34139.1 GBHO01002235 JAG41369.1 GGFJ01002681 MBW51822.1 GGFJ01002663 MBW51804.1 AAAB01007181 EAA02909.4 AJVK01013553 AJVK01013554 AAAB01008966 EAA13010.4 GDHC01016579 JAQ02050.1 KA644885 AFP59514.1 GBHO01002234 JAG41370.1 GGFL01003132 MBW67310.1 GGFL01003107 MBW67285.1 GGFL01003106 MBW67284.1 ADMH02001789 ETN61084.1 AXCM01002708 AXCN02001123 APCN01004446 APCN01004447 CH964278 EDW85369.2 KK854125 PTY11742.1 ATLV01027105 KE525423 KFB53964.1 MRZV01000061 PIK60273.1 BX119991 BX957355 JARO02000522 KPP78276.1 KY652752 ATU31576.1 AB987999 BAP46892.1 GGFM01006282 MBW27033.1 RJVU01036174 ROL47020.1
KQ459583 KPI98694.1 KQ461196 KPJ06457.1 BABH01010486 NWSH01006457 PCG63421.1 JTDY01000025 KOB79355.1 KK852820 KDR15587.1 NEVH01001351 PNF42922.1 GFTR01006180 JAW10246.1 UFQT01000416 SSX24119.1 PNF42925.1 GFDF01009200 JAV04884.1 CH933809 EDW19732.1 OUUW01000002 SPP76029.1 CM000159 EDW94547.1 CH940647 EDW70333.1 GFDF01009546 JAV04538.1 CH916366 EDV97137.1 CM000363 CM002912 EDX10491.1 KMY99645.1 CH954178 EDV51800.1 AE014296 BT030430 AAF49687.2 ABO52850.1 ABW08542.1 CH379069 EAL30690.2 CH479191 EDW26253.1 CH480815 EDW41521.1 CH477488 EAT40100.1 AJWK01014885 CH478204 EAT33567.1 GAKP01016896 GAKP01016895 JAC42057.1 GECU01025438 JAS82268.1 GEBQ01027228 JAT12749.1 DS232762 EDS45497.1 DS235771 EEB16764.1 KRF84861.1 GECZ01029884 GECZ01026048 GECZ01025808 GECZ01017396 GECZ01014368 GECZ01012569 GECZ01011268 GECZ01008724 GECZ01007339 JAS39885.1 JAS43721.1 JAS43961.1 JAS52373.1 JAS55401.1 JAS57200.1 JAS58501.1 JAS61045.1 JAS62430.1 GFDL01000105 JAV34940.1 JXUM01021127 KQ560592 KXJ81745.1 GFDL01000145 JAV34900.1 GFDL01000139 JAV34906.1 GFDL01000138 JAV34907.1 CP012525 ALC43818.1 GFDL01000125 JAV34920.1 CH902618 EDV40690.1 KRK01909.1 GFDL01000141 JAV34904.1 GAMC01010074 JAB96481.1 UFQT01002849 SSX34139.1 GBHO01002235 JAG41369.1 GGFJ01002681 MBW51822.1 GGFJ01002663 MBW51804.1 AAAB01007181 EAA02909.4 AJVK01013553 AJVK01013554 AAAB01008966 EAA13010.4 GDHC01016579 JAQ02050.1 KA644885 AFP59514.1 GBHO01002234 JAG41370.1 GGFL01003132 MBW67310.1 GGFL01003107 MBW67285.1 GGFL01003106 MBW67284.1 ADMH02001789 ETN61084.1 AXCM01002708 AXCN02001123 APCN01004446 APCN01004447 CH964278 EDW85369.2 KK854125 PTY11742.1 ATLV01027105 KE525423 KFB53964.1 MRZV01000061 PIK60273.1 BX119991 BX957355 JARO02000522 KPP78276.1 KY652752 ATU31576.1 AB987999 BAP46892.1 GGFM01006282 MBW27033.1 RJVU01036174 ROL47020.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000005204
UP000218220
UP000037510
+ More
UP000027135 UP000235965 UP000009192 UP000268350 UP000002282 UP000008792 UP000001070 UP000000304 UP000008711 UP000000803 UP000001819 UP000008744 UP000001292 UP000079169 UP000008820 UP000092461 UP000095300 UP000002320 UP000009046 UP000069940 UP000249989 UP000092553 UP000007801 UP000007062 UP000092462 UP000095301 UP000192221 UP000091820 UP000075881 UP000069272 UP000000673 UP000075880 UP000075883 UP000075886 UP000075840 UP000007798 UP000075884 UP000078200 UP000075920 UP000030765 UP000075885 UP000261440 UP000230750 UP000221080 UP000018467 UP000000437 UP000034805 UP000192224 UP000265140
UP000027135 UP000235965 UP000009192 UP000268350 UP000002282 UP000008792 UP000001070 UP000000304 UP000008711 UP000000803 UP000001819 UP000008744 UP000001292 UP000079169 UP000008820 UP000092461 UP000095300 UP000002320 UP000009046 UP000069940 UP000249989 UP000092553 UP000007801 UP000007062 UP000092462 UP000095301 UP000192221 UP000091820 UP000075881 UP000069272 UP000000673 UP000075880 UP000075883 UP000075886 UP000075840 UP000007798 UP000075884 UP000078200 UP000075920 UP000030765 UP000075885 UP000261440 UP000230750 UP000221080 UP000018467 UP000000437 UP000034805 UP000192224 UP000265140
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2H1X0G2
A0A2W1BQC4
A0A212EN13
A0A194Q129
A0A194QLQ2
H9J0Z3
+ More
A0A2A4IUM0 A0A0L7LV96 A0A067QYY1 A0A2J7RQ13 A0A224XCT1 A0A336M5G6 A0A2J7RQ10 A0A1L8DEX8 B4KXM5 A0A3B0JQU3 B4PIJ0 B4LFL5 A0A1L8DDM2 B4J2W7 B4QKI0 B3NHK5 A4IJ72 Q2M185 B4GUH0 B4HHV6 A0A3Q0J130 A0A1S4ED83 Q16ZM6 A0A1B0CJR3 Q16H51 A0A034VIU8 A0A1S4FIR7 A0A1B6I5Q9 A0A1I8PXS8 A0A1B6KMR7 B0XDI9 E0VTQ8 A0A0Q9WJC4 A0A1B6F1J0 A0A1Q3G554 A0A182GYE9 A0A1Q3G520 A0A1Q3G562 A0A1Q3G533 A0A0M5J5T8 A0A1Q3G539 B3M5Q6 A0A0R1DY55 A0A1Q3G531 W8BTE1 A0A336MYD4 A0A0A9ZAA5 A0A2M4BFL0 A0A2M4BFI7 Q7QLH2 A0A1B0GNR5 A0A1I8MYG1 Q7Q2X7 A0A146L5P5 T1P9P1 A0A0A9ZGZ4 A0A1S4H6I8 A0A2M4CPQ2 A0A1W4UMK0 A0A1A9WQ12 A0A182K1N3 A0A2M4CPU4 A0A2M4CPN0 A0A182FBS3 W5JE42 A0A182JFC6 A0A182LRW7 A0A182QJL7 A0A182HXZ9 B4NM01 A0A182N370 A0A1A9V5R9 A0A182W2C5 A0A2R7VWA6 A0A084WUS0 A0A182PAX6 A0A3B4EJM1 A0A2G8LJ35 A0A2D0PZX4 A0A3B1KKP0 A0A2R8Q6G1 F8W5D4 A0A0P7XSD3 A0A2D3E2E7 A0A3B1JPH6 A0A089ZW72 A0A2M3ZEV5 A0A1W5AQG3 A0A3P8Z3L9 A0A3N0YM80
A0A2A4IUM0 A0A0L7LV96 A0A067QYY1 A0A2J7RQ13 A0A224XCT1 A0A336M5G6 A0A2J7RQ10 A0A1L8DEX8 B4KXM5 A0A3B0JQU3 B4PIJ0 B4LFL5 A0A1L8DDM2 B4J2W7 B4QKI0 B3NHK5 A4IJ72 Q2M185 B4GUH0 B4HHV6 A0A3Q0J130 A0A1S4ED83 Q16ZM6 A0A1B0CJR3 Q16H51 A0A034VIU8 A0A1S4FIR7 A0A1B6I5Q9 A0A1I8PXS8 A0A1B6KMR7 B0XDI9 E0VTQ8 A0A0Q9WJC4 A0A1B6F1J0 A0A1Q3G554 A0A182GYE9 A0A1Q3G520 A0A1Q3G562 A0A1Q3G533 A0A0M5J5T8 A0A1Q3G539 B3M5Q6 A0A0R1DY55 A0A1Q3G531 W8BTE1 A0A336MYD4 A0A0A9ZAA5 A0A2M4BFL0 A0A2M4BFI7 Q7QLH2 A0A1B0GNR5 A0A1I8MYG1 Q7Q2X7 A0A146L5P5 T1P9P1 A0A0A9ZGZ4 A0A1S4H6I8 A0A2M4CPQ2 A0A1W4UMK0 A0A1A9WQ12 A0A182K1N3 A0A2M4CPU4 A0A2M4CPN0 A0A182FBS3 W5JE42 A0A182JFC6 A0A182LRW7 A0A182QJL7 A0A182HXZ9 B4NM01 A0A182N370 A0A1A9V5R9 A0A182W2C5 A0A2R7VWA6 A0A084WUS0 A0A182PAX6 A0A3B4EJM1 A0A2G8LJ35 A0A2D0PZX4 A0A3B1KKP0 A0A2R8Q6G1 F8W5D4 A0A0P7XSD3 A0A2D3E2E7 A0A3B1JPH6 A0A089ZW72 A0A2M3ZEV5 A0A1W5AQG3 A0A3P8Z3L9 A0A3N0YM80
PDB
4X82
E-value=2.18472e-05,
Score=115
Ontologies
GO
Topology
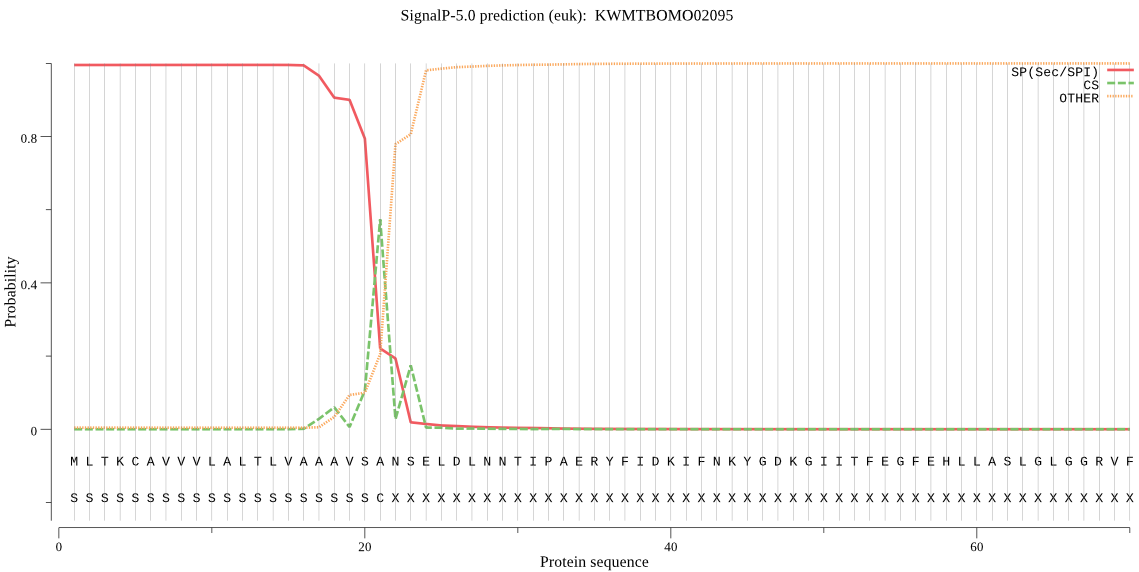
SignalP
Position: 1 - 21,
Likelihood: 0.995485
Length:
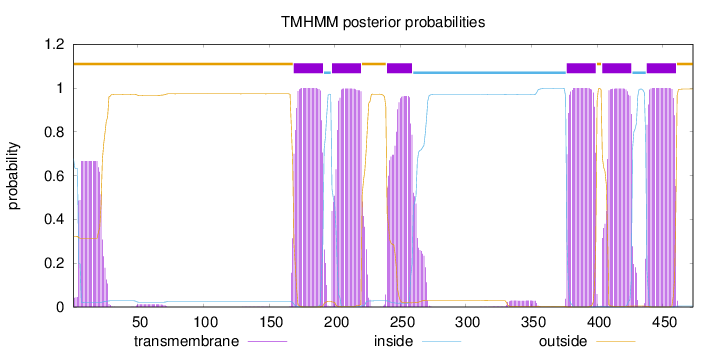
473
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
144.27305
Exp number, first 60 AAs:
12.65291
Total prob of N-in:
0.67742
POSSIBLE N-term signal
sequence
outside
1 - 168
TMhelix
169 - 191
inside
192 - 197
TMhelix
198 - 220
outside
221 - 239
TMhelix
240 - 259
inside
260 - 376
TMhelix
377 - 399
outside
400 - 403
TMhelix
404 - 426
inside
427 - 437
TMhelix
438 - 460
outside
461 - 473
Population Genetic Test Statistics
Pi
242.148534
Theta
192.560977
Tajima's D
1.145899
CLR
0.262787
CSRT
0.698565071746413
Interpretation
Uncertain
