Pre Gene Modal
BGIBMGA003182
Annotation
PREDICTED:_NADH_dehydrogenase_(ubiquinone)_complex_I?_assembly_factor_6_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.362 Mitochondrial Reliability : 1.473
Sequence
CDS
ATGGGTTTGGGTAGTAATATAATTAATTTTAAGACATACAAAATACTTAAAATGCCAGCATTTACTACTAGATTTCTCTCAAAAATAGCAGAGTCTAACCAGGACTCACTAAATTACTGTGCAAATTTAGTAAGGCAGCACGATTATGAAAATTTTTTGGCCACACTGCTCCTGACAAAGGCTGTAAGATCTCCGACGCTTGTAGTCAGAGCCTTCAATGTAGAAGTAGCTCGTATACAAGATCAAACCACCGATTCTCAGACAGCAGCTTTCAGATTGCAGTTCTGGAATGATACACTTAAACTACTCTATAAAAATGATCAAACTTTAAAAAATATTCCAGCAAACCCCATAGCACAAGAGCTGTTTAAGGTGTGTAACTCTTGCAAACTCCCAAAAAGATATTTAGAAAGATTAATTACAGCGAGAGGAAACATATTGAAATCAAAACACTTTCAAAGCTTGGAAGATTTAGAGAAATATGCCGAAGATTCAGTTAGTTCCATCTACTACCTCATTCTTAGTATTTCTGACGTTACCAATGTACATGCTGACCATGCCGCTTCACATTTGGGAAAGGCACAAGGAATTGTAAACATACTAAGATCAGTACATGTTGCTAGTTATCATAAAACGGTAACCTTACCGATGAATACGTTAATGAAATATGGGATCAGCCAAGAAAACGTTTTGAGAGGTATCGACAATGAGAATATGAGAAATGTTGCATTTGAAATAGCCACCAGAGCTAACAGTCATTTAGAAAAGGCTCAGGCAATTGATGTACCAAAAAAAACAAAGCAAATATTTTTACCTGCAGTAGCAGTAAACGCGTACTTGAAAAAACTACAAAAATGTAATTTTAACTTATATGACAAATCTTTACAATTGGGTAACTCTACTCTGCCACTAAATCTATATTACAATAGATTAATGAATAAATATTAA
Protein
MGLGSNIINFKTYKILKMPAFTTRFLSKIAESNQDSLNYCANLVRQHDYENFLATLLLTKAVRSPTLVVRAFNVEVARIQDQTTDSQTAAFRLQFWNDTLKLLYKNDQTLKNIPANPIAQELFKVCNSCKLPKRYLERLITARGNILKSKHFQSLEDLEKYAEDSVSSIYYLILSISDVTNVHADHAASHLGKAQGIVNILRSVHVASYHKTVTLPMNTLMKYGISQENVLRGIDNENMRNVAFEIATRANSHLEKAQAIDVPKKTKQIFLPAVAVNAYLKKLQKCNFNLYDKSLQLGNSTLPLNLYYNRLMNKY
Summary
Uniprot
A0A2A4JKP6
A0A2H1W0T0
A0A2W1BZM8
A0A212EN33
A0A2A4JKS0
A0A1E1WGT4
+ More
A0A194QNA8 A0A194Q074 A0A1B6E3J9 A0A1B6GRK5 A0A0P4VY62 R4FK79 A0A067QYX8 A0A182GV66 A0A182GQ33 E9FYX2 A0A0N7Z8V5 Q16X99 A0A0P5QLZ1 A0A0P5L111 A0A0P6IB09 A0A2J7R2E3 A0A1Y1K5Y6 K7IQ97 A0A232EX85 A0A2S2PLH9 A0A182KC83 A0A1J1I9A6 A0A0P5M030 A0A2H8TPH4 J9JYI3 A0A182VKN7 A0A0S7MI35 A0A3B3XEF7 A0A3P9PZJ0 A0A195E783 A0A2S2PY58 A0A182XBT5 A0A182MWH7 A0A3B3V7I6 Q7PS32 A0A2R2MP92 A0A182UDE4 A0A151I133 A0A182HMW9 B0W9G6 A0A182P8J4 U5ESL9 A0A182Q1J6 A0A336LQ12 T1IUF8 M4AGK9 A0A182SY24 E2A9L2 A0A158NG19 A0A087XB06 E2BN04 A0A146ZRC3 A0A0L0G8M4 A0A182N0H7 A0A2D0S159 F4X6K2 A0A131YTV9 A0A2D0S146 A0A1A8R664 A0A1A8MUG2 A0A182YK31 A0A224Z2V6 A0A1A8G210 A0A195F152 F1QP26 A0A0C9R1R5 A0A1A8AE03 A0A1A8KSF2 U4TZC1 A0A1A8BUC5 Q4V8Q6 A0A3B3CQ33 A0A0D2WP63 A0A3P9BPK9 A0A131YBA3 A0A1A7X550 A0A3P8NHA1 J3JUC7 A0A182J4S7 D6WN19 H2MKN3 A0A3P9IS88 A0A0L7R795 A0A146R0I6 A0A146LCH1 N6T7V9 I3IZ45 A0A3P9MGI4 A0A315VMZ9 A0A2K6GST9 B3N1D7
A0A194QNA8 A0A194Q074 A0A1B6E3J9 A0A1B6GRK5 A0A0P4VY62 R4FK79 A0A067QYX8 A0A182GV66 A0A182GQ33 E9FYX2 A0A0N7Z8V5 Q16X99 A0A0P5QLZ1 A0A0P5L111 A0A0P6IB09 A0A2J7R2E3 A0A1Y1K5Y6 K7IQ97 A0A232EX85 A0A2S2PLH9 A0A182KC83 A0A1J1I9A6 A0A0P5M030 A0A2H8TPH4 J9JYI3 A0A182VKN7 A0A0S7MI35 A0A3B3XEF7 A0A3P9PZJ0 A0A195E783 A0A2S2PY58 A0A182XBT5 A0A182MWH7 A0A3B3V7I6 Q7PS32 A0A2R2MP92 A0A182UDE4 A0A151I133 A0A182HMW9 B0W9G6 A0A182P8J4 U5ESL9 A0A182Q1J6 A0A336LQ12 T1IUF8 M4AGK9 A0A182SY24 E2A9L2 A0A158NG19 A0A087XB06 E2BN04 A0A146ZRC3 A0A0L0G8M4 A0A182N0H7 A0A2D0S159 F4X6K2 A0A131YTV9 A0A2D0S146 A0A1A8R664 A0A1A8MUG2 A0A182YK31 A0A224Z2V6 A0A1A8G210 A0A195F152 F1QP26 A0A0C9R1R5 A0A1A8AE03 A0A1A8KSF2 U4TZC1 A0A1A8BUC5 Q4V8Q6 A0A3B3CQ33 A0A0D2WP63 A0A3P9BPK9 A0A131YBA3 A0A1A7X550 A0A3P8NHA1 J3JUC7 A0A182J4S7 D6WN19 H2MKN3 A0A3P9IS88 A0A0L7R795 A0A146R0I6 A0A146LCH1 N6T7V9 I3IZ45 A0A3P9MGI4 A0A315VMZ9 A0A2K6GST9 B3N1D7
Pubmed
EMBL
NWSH01001239
PCG72003.1
ODYU01005642
SOQ46700.1
KZ149933
PZC77283.1
+ More
AGBW02013744 OWR42888.1 PCG72002.1 GDQN01004862 JAT86192.1 KQ461196 KPJ06460.1 KQ459583 KPI98698.1 GEDC01004829 JAS32469.1 GECZ01004705 JAS65064.1 GDRN01088085 JAI60921.1 ACPB03007690 GAHY01001983 JAA75527.1 KK852820 KDR15582.1 JXUM01090419 KQ563838 KXJ73195.1 JXUM01079645 KQ563138 KXJ74414.1 GL732527 EFX87698.1 GDKW01002291 JAI54304.1 CH477545 EAT39219.1 GDIP01223743 GDIQ01133045 JAI99658.1 JAL18681.1 GDIQ01185279 JAK66446.1 GDIQ01008791 JAN85946.1 NEVH01007831 PNF35010.1 GEZM01094181 JAV55838.1 NNAY01001780 OXU22939.1 GGMR01017656 MBY30275.1 CVRI01000044 CRK96863.1 GDIQ01171687 JAK80038.1 GFXV01004240 MBW16045.1 ABLF02019952 GBYX01021311 JAP01403.1 KQ979568 KYN20956.1 GGMS01001156 MBY70359.1 AXCM01002316 AAAB01008846 EAA06250.6 KQ976580 KYM79898.1 APCN01004922 DS231864 EDS40191.1 GANO01003187 JAB56684.1 AXCN02001411 UFQT01000107 SSX20092.1 JH431533 GL437917 EFN69902.1 ADTU01014889 AYCK01014953 GL449382 EFN82908.1 GCES01017440 JAR68883.1 KQ241706 KNC85360.1 GL888818 EGI57962.1 GEDV01007216 JAP81341.1 HAEH01014854 SBS01187.1 HAEF01019133 SBR60292.1 GFPF01010125 MAA21271.1 HAEB01017877 SBQ64404.1 KQ981864 KYN34193.1 BX294187 GBYB01000786 JAG70553.1 HADY01013970 HAEJ01017996 SBP52455.1 HAED01021241 HAEE01015098 SBR35148.1 KB631899 ERL86959.1 HADZ01006472 HAEA01013022 SBP70413.1 BC097249 AAH97249.1 KE346363 KJE92383.1 GEFM01000034 JAP75762.1 HADW01011640 SBP13040.1 BT126840 AEE61802.1 KQ971343 EFA03248.1 KQ414642 KOC66745.1 GCES01124443 JAQ61879.1 GDHC01013280 JAQ05349.1 APGK01040871 KB740984 ENN76314.1 AERX01041099 NHOQ01001373 PWA24923.1 CH902654 EDV33576.1
AGBW02013744 OWR42888.1 PCG72002.1 GDQN01004862 JAT86192.1 KQ461196 KPJ06460.1 KQ459583 KPI98698.1 GEDC01004829 JAS32469.1 GECZ01004705 JAS65064.1 GDRN01088085 JAI60921.1 ACPB03007690 GAHY01001983 JAA75527.1 KK852820 KDR15582.1 JXUM01090419 KQ563838 KXJ73195.1 JXUM01079645 KQ563138 KXJ74414.1 GL732527 EFX87698.1 GDKW01002291 JAI54304.1 CH477545 EAT39219.1 GDIP01223743 GDIQ01133045 JAI99658.1 JAL18681.1 GDIQ01185279 JAK66446.1 GDIQ01008791 JAN85946.1 NEVH01007831 PNF35010.1 GEZM01094181 JAV55838.1 NNAY01001780 OXU22939.1 GGMR01017656 MBY30275.1 CVRI01000044 CRK96863.1 GDIQ01171687 JAK80038.1 GFXV01004240 MBW16045.1 ABLF02019952 GBYX01021311 JAP01403.1 KQ979568 KYN20956.1 GGMS01001156 MBY70359.1 AXCM01002316 AAAB01008846 EAA06250.6 KQ976580 KYM79898.1 APCN01004922 DS231864 EDS40191.1 GANO01003187 JAB56684.1 AXCN02001411 UFQT01000107 SSX20092.1 JH431533 GL437917 EFN69902.1 ADTU01014889 AYCK01014953 GL449382 EFN82908.1 GCES01017440 JAR68883.1 KQ241706 KNC85360.1 GL888818 EGI57962.1 GEDV01007216 JAP81341.1 HAEH01014854 SBS01187.1 HAEF01019133 SBR60292.1 GFPF01010125 MAA21271.1 HAEB01017877 SBQ64404.1 KQ981864 KYN34193.1 BX294187 GBYB01000786 JAG70553.1 HADY01013970 HAEJ01017996 SBP52455.1 HAED01021241 HAEE01015098 SBR35148.1 KB631899 ERL86959.1 HADZ01006472 HAEA01013022 SBP70413.1 BC097249 AAH97249.1 KE346363 KJE92383.1 GEFM01000034 JAP75762.1 HADW01011640 SBP13040.1 BT126840 AEE61802.1 KQ971343 EFA03248.1 KQ414642 KOC66745.1 GCES01124443 JAQ61879.1 GDHC01013280 JAQ05349.1 APGK01040871 KB740984 ENN76314.1 AERX01041099 NHOQ01001373 PWA24923.1 CH902654 EDV33576.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000015103
UP000027135
+ More
UP000069940 UP000249989 UP000000305 UP000008820 UP000235965 UP000002358 UP000215335 UP000075881 UP000183832 UP000007819 UP000075903 UP000261480 UP000242638 UP000078492 UP000076407 UP000075883 UP000261500 UP000007062 UP000085678 UP000075902 UP000078540 UP000075840 UP000002320 UP000075885 UP000075886 UP000002852 UP000075901 UP000000311 UP000005205 UP000028760 UP000008237 UP000054560 UP000075884 UP000221080 UP000007755 UP000076408 UP000078541 UP000000437 UP000030742 UP000261560 UP000008743 UP000265160 UP000265100 UP000075880 UP000007266 UP000001038 UP000265200 UP000053825 UP000019118 UP000005207 UP000265180 UP000233160 UP000007801
UP000069940 UP000249989 UP000000305 UP000008820 UP000235965 UP000002358 UP000215335 UP000075881 UP000183832 UP000007819 UP000075903 UP000261480 UP000242638 UP000078492 UP000076407 UP000075883 UP000261500 UP000007062 UP000085678 UP000075902 UP000078540 UP000075840 UP000002320 UP000075885 UP000075886 UP000002852 UP000075901 UP000000311 UP000005205 UP000028760 UP000008237 UP000054560 UP000075884 UP000221080 UP000007755 UP000076408 UP000078541 UP000000437 UP000030742 UP000261560 UP000008743 UP000265160 UP000265100 UP000075880 UP000007266 UP000001038 UP000265200 UP000053825 UP000019118 UP000005207 UP000265180 UP000233160 UP000007801
Interpro
IPR008949
Isoprenoid_synthase_dom_sf
SUPFAM
SSF48576
SSF48576
Gene 3D
ProteinModelPortal
A0A2A4JKP6
A0A2H1W0T0
A0A2W1BZM8
A0A212EN33
A0A2A4JKS0
A0A1E1WGT4
+ More
A0A194QNA8 A0A194Q074 A0A1B6E3J9 A0A1B6GRK5 A0A0P4VY62 R4FK79 A0A067QYX8 A0A182GV66 A0A182GQ33 E9FYX2 A0A0N7Z8V5 Q16X99 A0A0P5QLZ1 A0A0P5L111 A0A0P6IB09 A0A2J7R2E3 A0A1Y1K5Y6 K7IQ97 A0A232EX85 A0A2S2PLH9 A0A182KC83 A0A1J1I9A6 A0A0P5M030 A0A2H8TPH4 J9JYI3 A0A182VKN7 A0A0S7MI35 A0A3B3XEF7 A0A3P9PZJ0 A0A195E783 A0A2S2PY58 A0A182XBT5 A0A182MWH7 A0A3B3V7I6 Q7PS32 A0A2R2MP92 A0A182UDE4 A0A151I133 A0A182HMW9 B0W9G6 A0A182P8J4 U5ESL9 A0A182Q1J6 A0A336LQ12 T1IUF8 M4AGK9 A0A182SY24 E2A9L2 A0A158NG19 A0A087XB06 E2BN04 A0A146ZRC3 A0A0L0G8M4 A0A182N0H7 A0A2D0S159 F4X6K2 A0A131YTV9 A0A2D0S146 A0A1A8R664 A0A1A8MUG2 A0A182YK31 A0A224Z2V6 A0A1A8G210 A0A195F152 F1QP26 A0A0C9R1R5 A0A1A8AE03 A0A1A8KSF2 U4TZC1 A0A1A8BUC5 Q4V8Q6 A0A3B3CQ33 A0A0D2WP63 A0A3P9BPK9 A0A131YBA3 A0A1A7X550 A0A3P8NHA1 J3JUC7 A0A182J4S7 D6WN19 H2MKN3 A0A3P9IS88 A0A0L7R795 A0A146R0I6 A0A146LCH1 N6T7V9 I3IZ45 A0A3P9MGI4 A0A315VMZ9 A0A2K6GST9 B3N1D7
A0A194QNA8 A0A194Q074 A0A1B6E3J9 A0A1B6GRK5 A0A0P4VY62 R4FK79 A0A067QYX8 A0A182GV66 A0A182GQ33 E9FYX2 A0A0N7Z8V5 Q16X99 A0A0P5QLZ1 A0A0P5L111 A0A0P6IB09 A0A2J7R2E3 A0A1Y1K5Y6 K7IQ97 A0A232EX85 A0A2S2PLH9 A0A182KC83 A0A1J1I9A6 A0A0P5M030 A0A2H8TPH4 J9JYI3 A0A182VKN7 A0A0S7MI35 A0A3B3XEF7 A0A3P9PZJ0 A0A195E783 A0A2S2PY58 A0A182XBT5 A0A182MWH7 A0A3B3V7I6 Q7PS32 A0A2R2MP92 A0A182UDE4 A0A151I133 A0A182HMW9 B0W9G6 A0A182P8J4 U5ESL9 A0A182Q1J6 A0A336LQ12 T1IUF8 M4AGK9 A0A182SY24 E2A9L2 A0A158NG19 A0A087XB06 E2BN04 A0A146ZRC3 A0A0L0G8M4 A0A182N0H7 A0A2D0S159 F4X6K2 A0A131YTV9 A0A2D0S146 A0A1A8R664 A0A1A8MUG2 A0A182YK31 A0A224Z2V6 A0A1A8G210 A0A195F152 F1QP26 A0A0C9R1R5 A0A1A8AE03 A0A1A8KSF2 U4TZC1 A0A1A8BUC5 Q4V8Q6 A0A3B3CQ33 A0A0D2WP63 A0A3P9BPK9 A0A131YBA3 A0A1A7X550 A0A3P8NHA1 J3JUC7 A0A182J4S7 D6WN19 H2MKN3 A0A3P9IS88 A0A0L7R795 A0A146R0I6 A0A146LCH1 N6T7V9 I3IZ45 A0A3P9MGI4 A0A315VMZ9 A0A2K6GST9 B3N1D7
Ontologies
GO
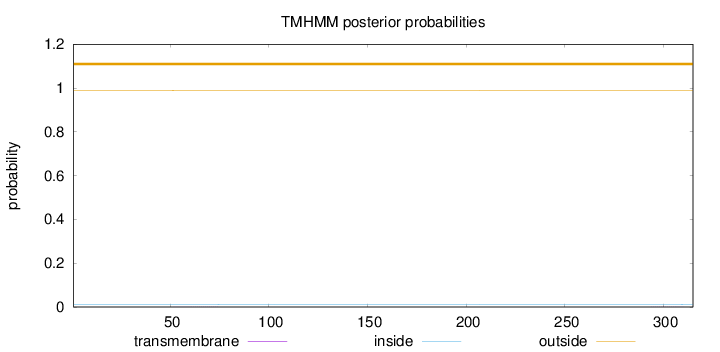
Topology
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01071
Exp number, first 60 AAs:
0.00261
Total prob of N-in:
0.01195
outside
1 - 315
Population Genetic Test Statistics
Pi
274.345457
Theta
211.005662
Tajima's D
0.110203
CLR
127.694687
CSRT
0.398230088495575
Interpretation
Possibly Positive selection
