Gene
KWMTBOMO02082 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002981
Annotation
profilin_[Bombyx_mori]
Full name
Profilin
Alternative Name
Protein chickadee
Location in the cell
Cytoplasmic Reliability : 2.2
Sequence
CDS
ATGAGCTGGCAAGATTATGTCGACAAACAGTTAATGGCATCTAGATGTGTCACAAAAGCTGCCATTGCCGGTCATGATGGCAATGTGTGGGCAAAGTCGGAAGGCTTCGAAATTTCAAAAGATGAAGTGGCGAAGATTGTGGCTGGCTTTGAGAATGAATCACTGCTAACGAGTGGCGGCGTGACGATAGCGGGCACGCGGTACATCTACCTCAGTGGCACAGACCATATCATCCGCGCGAAGCTTGGCAAGGTCGGCGTGCATTGCATGAAGACACAGCAAGCTGTGGTCATTTCTCTCTATGAAGAACCCATCCAACCTCAGCAGGCCGCATCTGTCGTGGAGAAGTTAGGAGAATATTTAATTACCTGTGGTTATTAG
Protein
MSWQDYVDKQLMASRCVTKAAIAGHDGNVWAKSEGFEISKDEVAKIVAGFENESLLTSGGVTIAGTRYIYLSGTDHIIRAKLGKVGVHCMKTQQAVVISLYEEPIQPQQAASVVEKLGEYLITCGY
Summary
Description
Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations.
Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations. By binding to PIP2, it inhibits the formation of IP3 and DG (By similarity).
Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations. By binding to PIP2, it inhibits the formation of IP3 and DG (By similarity).
Subunit
Occurs in many kinds of cells as a complex with monomeric actin in a 1:1 ratio.
Similarity
Belongs to the profilin family.
Keywords
Actin-binding
Complete proteome
Cytoplasm
Cytoskeleton
Reference proteome
Feature
chain Profilin
Uniprot
A1E8I8
Q68HB4
A0A0L7LKM4
I7G0X7
A0A1E1WDV7
A0A2A4JYL4
+ More
I4DRG9 A0A222AJF8 A0A212FD41 S4P8I6 A0A067QYW4 A0A0A9WSP9 A0A0U4B5D9 A0A2R7WJM6 R4WQQ2 J3JTV4 A0A139WI06 J3JX68 A0A195AVA9 A0A195ETL5 A0A158NHK5 A0A1B6LUA2 A0A1B6GYU7 E2APZ9 A0A2P8YC67 A0A1B6ITY7 A0A195C3G6 A0A1Y1MU09 A0A026WB74 A0A1W4XKU8 A0A146KJP4 A0A2A3EQJ2 V9IGT1 A0A087ZNT2 A0A232EQC9 K7IQ05 V5GLW9 A0A224Y093 A0A0V0G4A7 A0A023F948 A0A069DNN0 A0A154PS72 A0A0L7RAG9 Q5TP19 A0A310SRH1 A0A1D1XF39 T1DFI4 A0A1Q3F8G0 B0W2U6 T1DQL5 A0A2M3Z0V6 A0A2M4A439 W5JBU9 A0A023EFI4 A0A0P4VU02 R4G508 Q6QEJ7 Q16JE9 A0A1B6DCQ5 A0A0T6AWK4 E0VF14 A0A1J1I5Z8 T1PA74 A0A0C9RAU5 A0A1L8EHM5 A0A1L8E5E0 U5EZ71 A0A0L0C5L6 A0A1D2NDF1 A0A1I8P5R4 W8CDG5 A0A0K8UUJ6 A0A0C9RGL0 A0A034VKC7 A0A0M3QT59 A0A1A9Y520 A0A1A9UYB4 A0A1A9Z9G4 D3TQD6 B4MUZ7 B3MMQ1 B4LR73 A0A3B0KI43 Q29PL9 B4JC19 B4KGG2 D3TKT4 A0A1W4UBW4 A0A0P6IHW6 B3N5A1 A0A1I9WLF3 T1H7C2 B4I1N3 Q3YMV5 B4NZS9 M9NEU5 P25843 A0A0K8TNS2 A0A151WGI7 A0A336KD15
I4DRG9 A0A222AJF8 A0A212FD41 S4P8I6 A0A067QYW4 A0A0A9WSP9 A0A0U4B5D9 A0A2R7WJM6 R4WQQ2 J3JTV4 A0A139WI06 J3JX68 A0A195AVA9 A0A195ETL5 A0A158NHK5 A0A1B6LUA2 A0A1B6GYU7 E2APZ9 A0A2P8YC67 A0A1B6ITY7 A0A195C3G6 A0A1Y1MU09 A0A026WB74 A0A1W4XKU8 A0A146KJP4 A0A2A3EQJ2 V9IGT1 A0A087ZNT2 A0A232EQC9 K7IQ05 V5GLW9 A0A224Y093 A0A0V0G4A7 A0A023F948 A0A069DNN0 A0A154PS72 A0A0L7RAG9 Q5TP19 A0A310SRH1 A0A1D1XF39 T1DFI4 A0A1Q3F8G0 B0W2U6 T1DQL5 A0A2M3Z0V6 A0A2M4A439 W5JBU9 A0A023EFI4 A0A0P4VU02 R4G508 Q6QEJ7 Q16JE9 A0A1B6DCQ5 A0A0T6AWK4 E0VF14 A0A1J1I5Z8 T1PA74 A0A0C9RAU5 A0A1L8EHM5 A0A1L8E5E0 U5EZ71 A0A0L0C5L6 A0A1D2NDF1 A0A1I8P5R4 W8CDG5 A0A0K8UUJ6 A0A0C9RGL0 A0A034VKC7 A0A0M3QT59 A0A1A9Y520 A0A1A9UYB4 A0A1A9Z9G4 D3TQD6 B4MUZ7 B3MMQ1 B4LR73 A0A3B0KI43 Q29PL9 B4JC19 B4KGG2 D3TKT4 A0A1W4UBW4 A0A0P6IHW6 B3N5A1 A0A1I9WLF3 T1H7C2 B4I1N3 Q3YMV5 B4NZS9 M9NEU5 P25843 A0A0K8TNS2 A0A151WGI7 A0A336KD15
Pubmed
18633732
26227816
22651552
22118469
23622113
24845553
+ More
25401762 26823975 26694822 23691247 22516182 18362917 19820115 23537049 21347285 20798317 29403074 28004739 24508170 30249741 28648823 20075255 25474469 26334808 12364791 14747013 17210077 24330624 20920257 23761445 24945155 26483478 27129103 17510324 20566863 25315136 26108605 27289101 24495485 25348373 20353571 17994087 18057021 15632085 23185243 27538518 15917496 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 1339308 12537569 26369729
25401762 26823975 26694822 23691247 22516182 18362917 19820115 23537049 21347285 20798317 29403074 28004739 24508170 30249741 28648823 20075255 25474469 26334808 12364791 14747013 17210077 24330624 20920257 23761445 24945155 26483478 27129103 17510324 20566863 25315136 26108605 27289101 24495485 25348373 20353571 17994087 18057021 15632085 23185243 27538518 15917496 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 1339308 12537569 26369729
EMBL
EF112402
ABK97428.1
AY690617
JTDY01000719
KOB76103.1
JX087453
+ More
ODYU01008012 AFP36379.1 SOQ51264.1 GDQN01005896 JAT85158.1 NWSH01000373 PCG76866.1 AK405112 BAM20509.1 KY609793 ASO76491.1 AGBW02009105 OWR51666.1 GAIX01005911 JAA86649.1 KK852820 KDR15557.1 GBHO01035730 GBHO01035729 GBRD01001259 GBRD01001258 GDHC01001113 JAG07874.1 JAG07875.1 JAG64562.1 JAQ17516.1 KU218676 ALX00056.1 KK854931 PTY19758.1 AK417007 BAN20222.1 BT126663 AEE61626.1 KQ971342 KYB27628.1 APGK01037935 BT127836 KB740949 AEE62798.1 ENN77193.1 KQ976737 KYM75960.1 KQ981975 KYN31595.1 ADTU01015912 ADTU01015913 ADTU01015914 ADTU01015915 GEBQ01012719 JAT27258.1 GECZ01002182 JAS67587.1 GL441645 EFN64486.1 PYGN01000712 PSN41836.1 GECU01017324 JAS90382.1 KQ978350 KYM94731.1 GEZM01021258 JAV89049.1 KK107293 QOIP01000005 EZA53310.1 RLU22461.1 GDHC01021826 JAP96802.1 KZ288204 PBC33311.1 JR042933 AEY59691.1 NNAY01002818 OXU20506.1 GALX01003402 JAB65064.1 GFTR01002124 JAW14302.1 GECL01003333 JAP02791.1 GBBI01000701 JAC18011.1 GBGD01003474 JAC85415.1 KQ435090 KZC14597.1 KQ414619 KOC67761.1 AAAB01008980 EAL39221.3 KQ761343 OAD57879.1 GDJX01026916 JAT41020.1 GALA01000582 JAA94270.1 GFDL01011185 JAV23860.1 DS231829 EDS30305.1 GAMD01002409 JAA99181.1 GGFM01001399 MBW22150.1 GGFK01002190 MBW35511.1 ADMH02001847 GGFL01004245 ETN60893.1 MBW68423.1 JXUM01054759 JXUM01054760 JXUM01054761 JXUM01054762 GAPW01006018 GAPW01006017 JAC07581.1 GDKW01000568 JAI56027.1 ACPB03021270 GAHY01000905 JAA76605.1 AY545000 CH478012 EAT34394.1 GEDC01013866 GEDC01008505 JAS23432.1 JAS28793.1 LJIG01022646 KRT79464.1 DS235100 EEB11988.1 CVRI01000038 CRK94366.1 KA645040 KA649095 KA649185 AFP59669.1 GBYB01013604 JAG83371.1 GFDG01000706 JAV18093.1 GFDF01000117 JAV13967.1 GANO01001322 JAB58549.1 JRES01000864 KNC27688.1 LJIJ01000079 ODN03284.1 GAMC01001986 JAC04570.1 GDHF01025877 GDHF01022954 GDHF01022078 GDHF01011740 GDHF01004181 GDHF01000053 JAI26437.1 JAI29360.1 JAI30236.1 JAI40574.1 JAI48133.1 JAI52261.1 GBYB01006166 GBYB01006168 JAG75933.1 JAG75935.1 GAKP01016003 GAKP01016002 JAC42949.1 CP012523 ALC38343.1 CCAG010020379 EZ423638 ADD19914.1 CH963857 EDW76342.1 CH902620 EDV31942.2 KPU73710.1 CH940649 EDW63537.1 OUUW01000010 SPP86079.1 CH379058 EAL34274.1 KRT03334.1 CH916368 EDW03032.1 CH933807 EDW12158.1 KRG03107.1 EZ422021 ADD18444.1 GDIQ01004307 JAN90430.1 CH954177 EDV57931.1 KU932337 APA33973.1 CH480820 EDW54440.1 DQ062771 CM002910 AAY56644.1 KMY88420.1 CM000157 EDW88863.1 AE014134 KX531778 AFH03579.1 ANY27588.1 M84528 M84529 DQ062770 AY069444 AY128443 GDAI01001589 JAI16014.1 KQ983167 KYQ46950.1 UFQS01000281 UFQT01000281 SSX02379.1 SSX22754.1
ODYU01008012 AFP36379.1 SOQ51264.1 GDQN01005896 JAT85158.1 NWSH01000373 PCG76866.1 AK405112 BAM20509.1 KY609793 ASO76491.1 AGBW02009105 OWR51666.1 GAIX01005911 JAA86649.1 KK852820 KDR15557.1 GBHO01035730 GBHO01035729 GBRD01001259 GBRD01001258 GDHC01001113 JAG07874.1 JAG07875.1 JAG64562.1 JAQ17516.1 KU218676 ALX00056.1 KK854931 PTY19758.1 AK417007 BAN20222.1 BT126663 AEE61626.1 KQ971342 KYB27628.1 APGK01037935 BT127836 KB740949 AEE62798.1 ENN77193.1 KQ976737 KYM75960.1 KQ981975 KYN31595.1 ADTU01015912 ADTU01015913 ADTU01015914 ADTU01015915 GEBQ01012719 JAT27258.1 GECZ01002182 JAS67587.1 GL441645 EFN64486.1 PYGN01000712 PSN41836.1 GECU01017324 JAS90382.1 KQ978350 KYM94731.1 GEZM01021258 JAV89049.1 KK107293 QOIP01000005 EZA53310.1 RLU22461.1 GDHC01021826 JAP96802.1 KZ288204 PBC33311.1 JR042933 AEY59691.1 NNAY01002818 OXU20506.1 GALX01003402 JAB65064.1 GFTR01002124 JAW14302.1 GECL01003333 JAP02791.1 GBBI01000701 JAC18011.1 GBGD01003474 JAC85415.1 KQ435090 KZC14597.1 KQ414619 KOC67761.1 AAAB01008980 EAL39221.3 KQ761343 OAD57879.1 GDJX01026916 JAT41020.1 GALA01000582 JAA94270.1 GFDL01011185 JAV23860.1 DS231829 EDS30305.1 GAMD01002409 JAA99181.1 GGFM01001399 MBW22150.1 GGFK01002190 MBW35511.1 ADMH02001847 GGFL01004245 ETN60893.1 MBW68423.1 JXUM01054759 JXUM01054760 JXUM01054761 JXUM01054762 GAPW01006018 GAPW01006017 JAC07581.1 GDKW01000568 JAI56027.1 ACPB03021270 GAHY01000905 JAA76605.1 AY545000 CH478012 EAT34394.1 GEDC01013866 GEDC01008505 JAS23432.1 JAS28793.1 LJIG01022646 KRT79464.1 DS235100 EEB11988.1 CVRI01000038 CRK94366.1 KA645040 KA649095 KA649185 AFP59669.1 GBYB01013604 JAG83371.1 GFDG01000706 JAV18093.1 GFDF01000117 JAV13967.1 GANO01001322 JAB58549.1 JRES01000864 KNC27688.1 LJIJ01000079 ODN03284.1 GAMC01001986 JAC04570.1 GDHF01025877 GDHF01022954 GDHF01022078 GDHF01011740 GDHF01004181 GDHF01000053 JAI26437.1 JAI29360.1 JAI30236.1 JAI40574.1 JAI48133.1 JAI52261.1 GBYB01006166 GBYB01006168 JAG75933.1 JAG75935.1 GAKP01016003 GAKP01016002 JAC42949.1 CP012523 ALC38343.1 CCAG010020379 EZ423638 ADD19914.1 CH963857 EDW76342.1 CH902620 EDV31942.2 KPU73710.1 CH940649 EDW63537.1 OUUW01000010 SPP86079.1 CH379058 EAL34274.1 KRT03334.1 CH916368 EDW03032.1 CH933807 EDW12158.1 KRG03107.1 EZ422021 ADD18444.1 GDIQ01004307 JAN90430.1 CH954177 EDV57931.1 KU932337 APA33973.1 CH480820 EDW54440.1 DQ062771 CM002910 AAY56644.1 KMY88420.1 CM000157 EDW88863.1 AE014134 KX531778 AFH03579.1 ANY27588.1 M84528 M84529 DQ062770 AY069444 AY128443 GDAI01001589 JAI16014.1 KQ983167 KYQ46950.1 UFQS01000281 UFQT01000281 SSX02379.1 SSX22754.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000027135
UP000007266
+ More
UP000019118 UP000078540 UP000078541 UP000005205 UP000000311 UP000245037 UP000078542 UP000053097 UP000279307 UP000192223 UP000242457 UP000005203 UP000215335 UP000002358 UP000076502 UP000053825 UP000007062 UP000002320 UP000000673 UP000069940 UP000015103 UP000008820 UP000009046 UP000183832 UP000095301 UP000037069 UP000094527 UP000095300 UP000092553 UP000092443 UP000078200 UP000092445 UP000092444 UP000007798 UP000007801 UP000008792 UP000268350 UP000001819 UP000001070 UP000009192 UP000192221 UP000008711 UP000015102 UP000001292 UP000002282 UP000000803 UP000075809
UP000019118 UP000078540 UP000078541 UP000005205 UP000000311 UP000245037 UP000078542 UP000053097 UP000279307 UP000192223 UP000242457 UP000005203 UP000215335 UP000002358 UP000076502 UP000053825 UP000007062 UP000002320 UP000000673 UP000069940 UP000015103 UP000008820 UP000009046 UP000183832 UP000095301 UP000037069 UP000094527 UP000095300 UP000092553 UP000092443 UP000078200 UP000092445 UP000092444 UP000007798 UP000007801 UP000008792 UP000268350 UP000001819 UP000001070 UP000009192 UP000192221 UP000008711 UP000015102 UP000001292 UP000002282 UP000000803 UP000075809
PRIDE
Pfam
PF00235 Profilin
SUPFAM
SSF55770
SSF55770
CDD
ProteinModelPortal
A1E8I8
Q68HB4
A0A0L7LKM4
I7G0X7
A0A1E1WDV7
A0A2A4JYL4
+ More
I4DRG9 A0A222AJF8 A0A212FD41 S4P8I6 A0A067QYW4 A0A0A9WSP9 A0A0U4B5D9 A0A2R7WJM6 R4WQQ2 J3JTV4 A0A139WI06 J3JX68 A0A195AVA9 A0A195ETL5 A0A158NHK5 A0A1B6LUA2 A0A1B6GYU7 E2APZ9 A0A2P8YC67 A0A1B6ITY7 A0A195C3G6 A0A1Y1MU09 A0A026WB74 A0A1W4XKU8 A0A146KJP4 A0A2A3EQJ2 V9IGT1 A0A087ZNT2 A0A232EQC9 K7IQ05 V5GLW9 A0A224Y093 A0A0V0G4A7 A0A023F948 A0A069DNN0 A0A154PS72 A0A0L7RAG9 Q5TP19 A0A310SRH1 A0A1D1XF39 T1DFI4 A0A1Q3F8G0 B0W2U6 T1DQL5 A0A2M3Z0V6 A0A2M4A439 W5JBU9 A0A023EFI4 A0A0P4VU02 R4G508 Q6QEJ7 Q16JE9 A0A1B6DCQ5 A0A0T6AWK4 E0VF14 A0A1J1I5Z8 T1PA74 A0A0C9RAU5 A0A1L8EHM5 A0A1L8E5E0 U5EZ71 A0A0L0C5L6 A0A1D2NDF1 A0A1I8P5R4 W8CDG5 A0A0K8UUJ6 A0A0C9RGL0 A0A034VKC7 A0A0M3QT59 A0A1A9Y520 A0A1A9UYB4 A0A1A9Z9G4 D3TQD6 B4MUZ7 B3MMQ1 B4LR73 A0A3B0KI43 Q29PL9 B4JC19 B4KGG2 D3TKT4 A0A1W4UBW4 A0A0P6IHW6 B3N5A1 A0A1I9WLF3 T1H7C2 B4I1N3 Q3YMV5 B4NZS9 M9NEU5 P25843 A0A0K8TNS2 A0A151WGI7 A0A336KD15
I4DRG9 A0A222AJF8 A0A212FD41 S4P8I6 A0A067QYW4 A0A0A9WSP9 A0A0U4B5D9 A0A2R7WJM6 R4WQQ2 J3JTV4 A0A139WI06 J3JX68 A0A195AVA9 A0A195ETL5 A0A158NHK5 A0A1B6LUA2 A0A1B6GYU7 E2APZ9 A0A2P8YC67 A0A1B6ITY7 A0A195C3G6 A0A1Y1MU09 A0A026WB74 A0A1W4XKU8 A0A146KJP4 A0A2A3EQJ2 V9IGT1 A0A087ZNT2 A0A232EQC9 K7IQ05 V5GLW9 A0A224Y093 A0A0V0G4A7 A0A023F948 A0A069DNN0 A0A154PS72 A0A0L7RAG9 Q5TP19 A0A310SRH1 A0A1D1XF39 T1DFI4 A0A1Q3F8G0 B0W2U6 T1DQL5 A0A2M3Z0V6 A0A2M4A439 W5JBU9 A0A023EFI4 A0A0P4VU02 R4G508 Q6QEJ7 Q16JE9 A0A1B6DCQ5 A0A0T6AWK4 E0VF14 A0A1J1I5Z8 T1PA74 A0A0C9RAU5 A0A1L8EHM5 A0A1L8E5E0 U5EZ71 A0A0L0C5L6 A0A1D2NDF1 A0A1I8P5R4 W8CDG5 A0A0K8UUJ6 A0A0C9RGL0 A0A034VKC7 A0A0M3QT59 A0A1A9Y520 A0A1A9UYB4 A0A1A9Z9G4 D3TQD6 B4MUZ7 B3MMQ1 B4LR73 A0A3B0KI43 Q29PL9 B4JC19 B4KGG2 D3TKT4 A0A1W4UBW4 A0A0P6IHW6 B3N5A1 A0A1I9WLF3 T1H7C2 B4I1N3 Q3YMV5 B4NZS9 M9NEU5 P25843 A0A0K8TNS2 A0A151WGI7 A0A336KD15
PDB
2ACG
E-value=4.76859e-25,
Score=277
Ontologies
GO
GO:0005856
GO:0003779
GO:0005737
GO:0042989
GO:0003785
GO:0005938
GO:0035019
GO:0007488
GO:0032507
GO:0007015
GO:0007436
GO:0007420
GO:0048842
GO:0016322
GO:0048471
GO:0000281
GO:0042060
GO:0048680
GO:0007411
GO:0043005
GO:0030717
GO:0035193
GO:0051491
GO:0036098
GO:0045451
GO:0000915
GO:0000902
GO:0007300
GO:0030041
GO:0007391
GO:0005524
GO:0004842
GO:0016567
GO:0046872
GO:0003676
GO:0005739
GO:0006635
GO:0016507
GO:0048384
PANTHER
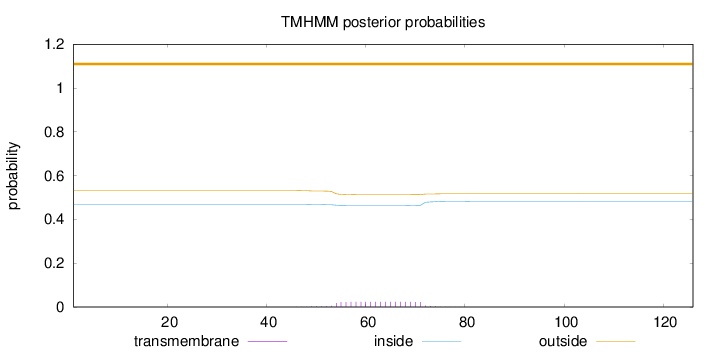
Topology
Subcellular location
Length:
126
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44543
Exp number, first 60 AAs:
0.17511
Total prob of N-in:
0.46832
outside
1 - 126
Population Genetic Test Statistics
Pi
254.900596
Theta
196.810306
Tajima's D
0.922549
CLR
0
CSRT
0.639118044097795
Interpretation
Uncertain
