Gene
KWMTBOMO02081 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003186
Annotation
PREDICTED:_eukaryotic_translation_initiation_factor_4A_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.849 Nuclear Reliability : 2.48
Sequence
CDS
ATGTCTTATTCGTCTGAAAGAAGGTCAGAAGATTGGCCAGAGGATTCCAAAAATGGGCCATCGAAGGATCAAGGTAGTTACGATGGACCTCCGGGAATGGACCCTGGGACACTTGACACCGACTGGGATCAAGTTGTCGAAACCTTCGATGACATGAACCTCAAAGAAGAATTGTTGAGAGGCATATACGCCTATGGTTTTGAAAAACCTTCTGCAATCCAGCAACGCGCAATAATGCCTTGCATCCAAGGACGCGATGTTATCGCTCAAGCCCAGTCAGGAACTGGAAAAACTGCTACTTTCTCTATATCGATTCTACAACAAATCGATACAAGCATTCGTGAATGTCAAGCTTTGATCCTGGCTCCCACAAGAGAGCTGGCCCAACAAATTCAGAAGGTGGTGATAGCTCTTGGTGATCACTTGAATGCTAAATGCCATGCTTGCATTGGTGGCACCAATGTCCGTGAAGATATTCGCCAACTGGAGAGTGGTGTTCATGTGGTGGTGGGCACTCCAGGTCGTGTATATGATATGATAACTCGTCGTGCGCTTCATGCCAACACCATCAAACTTTTTGTTCTTGATGAAGCTGATGAAATGTTGTCCAGAGGTTTCAAGGACCAGATCCATGATGTCTTCAAGATGTTGTCAGCTGATGTTCAAGTCATATTACTCTCTGCTACCATGCCTGATGATGTATTGGAAGTATCTCGATGCTTTATGAGAGATCCTGTACGCATACTTGTACAGAAGGAAGAGCTTACCCTGGAAGGTATTAAACAATTTTACATTGCAATTGAATTAGAAGAATGGAAGCTGGAAACTCTGTGTGACCTGTATGATACACTGTCTATTGCACAAGCTGTAATTTTCTGCAACACCCGTCGCAAGGTGGATTGGCTCACTGAATCTATGCATCTGCGTGACTTTACTGTATCTGCTATGCATGGAGACATGGATCAACGTGAGCGTGAAGTGATCATGAGGCAGTTTCGTACTGGCTCTTCTCGTGTCTTGATCACCACTGATTTATTGGCACGTGGTATTGATGTACAGCAAGTTTCCTGCGTCATCAACTATGATCTGCCATCCAACCGTGAAAATTATATTCACAGGATTGGACGAGGTGGACGTTTTGGTCGTAAGGGAATTGCTATCAACTTTGTGACTGAAGCTGACAGGAGAGCACTGAAGGATATTGAGGACTTCTACCACACTAGTATCGTTGAAATGCCCAGTGATGTGGCCAACCTCATCTAA
Protein
MSYSSERRSEDWPEDSKNGPSKDQGSYDGPPGMDPGTLDTDWDQVVETFDDMNLKEELLRGIYAYGFEKPSAIQQRAIMPCIQGRDVIAQAQSGTGKTATFSISILQQIDTSIRECQALILAPTRELAQQIQKVVIALGDHLNAKCHACIGGTNVREDIRQLESGVHVVVGTPGRVYDMITRRALHANTIKLFVLDEADEMLSRGFKDQIHDVFKMLSADVQVILLSATMPDDVLEVSRCFMRDPVRILVQKEELTLEGIKQFYIAIELEEWKLETLCDLYDTLSIAQAVIFCNTRRKVDWLTESMHLRDFTVSAMHGDMDQREREVIMRQFRTGSSRVLITTDLLARGIDVQQVSCVINYDLPSNRENYIHRIGRGGRFGRKGIAINFVTEADRRALKDIEDFYHTSIVEMPSDVANLI
Summary
Similarity
Belongs to the DEAD box helicase family.
Uniprot
Q1HPW2
Q285R3
A0A2A4JYK4
A0A2H1WDX4
A0A3G1T185
B0FYV2
+ More
N0ACL6 S4PHK3 A0A212FD75 I4DMD3 I4DIH2 A0A1B1LYS1 E1ZX82 F4WGJ4 A0A158NP28 A0A1W4XVZ8 A0A026WJQ4 E2C603 E9IEB5 A0A067R0Y2 A0A1B6F2F1 R4WDG0 A0A195F028 A0A151J9B0 A0A195BZM2 D6WKA5 A0A088AJJ6 V9ID91 A0A1D1Z3B1 A0A151IEQ7 A0A1B6J9R8 A0A1B6KUF3 A0A2J7PVH2 A0A2A3E853 A0A0C9QHZ8 A0A1Y1K7F5 A0A1B6EJ32 K4QDH4 V5GVE3 W8GNB0 A0A0T6AXY1 J3JTM5 K7J879 U4U468 A0A0L7REA5 A0A0M8ZQ94 A0A023FAM3 A0A154P5P5 A0A069DYY3 A0A1L8DH04 A0A023GJT3 G3MPG6 A0A023FZF6 A0A023FJS7 A0A224XR04 T1HS20 A0A1L8DH64 A0A131XEX0 A0A1L8DGZ6 A0A1E1X9E5 A0A224Z8H7 A0A131YS33 L7MAX7 A0A232EYW5 A0A2R5LN07 A0A151XDR0 A0A1B6C1X4 A0A0K8RLN8 A0A0C9RYG0 A0A0K8SSR9 A0A0A9ZJ75 A0A146M4N1 A0A087TAB2 A0A2L2YMJ4 B7P5C4 E9GA59 A0A0C9SCE0 A0A1W7R9F4 A0A0P5UMI5 A0A2H8TEY3 A0A131XZM9 A0A0N8BKN0 J9JQC9 A0A0P5UJ14 A0A0P4Z8M2 A0A0P5YFG9 A0A0P5KTM7 A0A0P5XGW5 A0A0P5QGP2 A0A0P5QRW3 A0A0P5GIX8 A0A0P5LJ14 A0A1B0CWZ5 A0A0J7KA78 A0A0P5SQT1 N6U958 A0A0P5I9K1 A0A0P5J364 A0A0N7ZZM5 A0A0P4WN89
N0ACL6 S4PHK3 A0A212FD75 I4DMD3 I4DIH2 A0A1B1LYS1 E1ZX82 F4WGJ4 A0A158NP28 A0A1W4XVZ8 A0A026WJQ4 E2C603 E9IEB5 A0A067R0Y2 A0A1B6F2F1 R4WDG0 A0A195F028 A0A151J9B0 A0A195BZM2 D6WKA5 A0A088AJJ6 V9ID91 A0A1D1Z3B1 A0A151IEQ7 A0A1B6J9R8 A0A1B6KUF3 A0A2J7PVH2 A0A2A3E853 A0A0C9QHZ8 A0A1Y1K7F5 A0A1B6EJ32 K4QDH4 V5GVE3 W8GNB0 A0A0T6AXY1 J3JTM5 K7J879 U4U468 A0A0L7REA5 A0A0M8ZQ94 A0A023FAM3 A0A154P5P5 A0A069DYY3 A0A1L8DH04 A0A023GJT3 G3MPG6 A0A023FZF6 A0A023FJS7 A0A224XR04 T1HS20 A0A1L8DH64 A0A131XEX0 A0A1L8DGZ6 A0A1E1X9E5 A0A224Z8H7 A0A131YS33 L7MAX7 A0A232EYW5 A0A2R5LN07 A0A151XDR0 A0A1B6C1X4 A0A0K8RLN8 A0A0C9RYG0 A0A0K8SSR9 A0A0A9ZJ75 A0A146M4N1 A0A087TAB2 A0A2L2YMJ4 B7P5C4 E9GA59 A0A0C9SCE0 A0A1W7R9F4 A0A0P5UMI5 A0A2H8TEY3 A0A131XZM9 A0A0N8BKN0 J9JQC9 A0A0P5UJ14 A0A0P4Z8M2 A0A0P5YFG9 A0A0P5KTM7 A0A0P5XGW5 A0A0P5QGP2 A0A0P5QRW3 A0A0P5GIX8 A0A0P5LJ14 A0A1B0CWZ5 A0A0J7KA78 A0A0P5SQT1 N6U958 A0A0P5I9K1 A0A0P5J364 A0A0N7ZZM5 A0A0P4WN89
Pubmed
19121390
23617895
23622113
22118469
22651552
26354079
+ More
27392023 20798317 21719571 21347285 24508170 30249741 21282665 24845553 23691247 18362917 19820115 28004739 22516182 20075255 23537049 25474469 26334808 22216098 28049606 28503490 28797301 26830274 25576852 28648823 25401762 26823975 26561354 21292972 26131772
27392023 20798317 21719571 21347285 24508170 30249741 21282665 24845553 23691247 18362917 19820115 28004739 22516182 20075255 23537049 25474469 26334808 22216098 28049606 28503490 28797301 26830274 25576852 28648823 25401762 26823975 26561354 21292972 26131772
EMBL
BABH01010502
DQ443290
ABF51379.1
DQ150250
AAZ80489.1
NWSH01000373
+ More
PCG76856.1 ODYU01008012 SOQ51263.1 MG846893 AXY94745.1 EU366906 ABY66383.1 KC481238 AGK45314.2 GAIX01003242 JAA89318.1 AGBW02009105 OWR51667.1 AK402451 BAM19073.1 AK401090 KQ459583 BAM17712.1 KPI98712.1 KU664392 ANS59102.1 GL435030 EFN74172.1 GL888137 EGI66659.1 ADTU01022136 ADTU01022137 KK107167 QOIP01000010 EZA56277.1 RLU17458.1 GL452895 EFN76608.1 GL762576 EFZ21079.1 KK852820 KDR15558.1 GECZ01025404 GECZ01012564 JAS44365.1 JAS57205.1 AK417653 BAN20868.1 KQ981891 KYN33828.1 KQ979425 KYN21607.1 KQ976394 KYM93358.1 KQ971342 EFA03968.2 JR039259 JR039260 AEY58606.1 AEY58607.1 GDJX01006590 GDJX01003127 JAT61346.1 JAT64809.1 KQ977858 KYM99329.1 GECU01011772 JAS95934.1 GEBQ01024898 JAT15079.1 NEVH01020961 PNF20334.1 KZ288349 PBC27386.1 GBYB01014298 JAG84065.1 GEZM01095793 GEZM01095792 JAV55346.1 GECZ01031842 GECZ01020215 JAS37927.1 JAS49554.1 HE962192 CCJ09440.1 GALX01004263 JAB64203.1 KF986613 AHK26791.1 LJIG01022646 KRT79463.1 BT126582 AEE61546.1 KB631923 ERL87143.1 KQ414612 KOC69173.1 KQ436240 KOX67369.1 GBBI01000195 GEMB01001334 JAC18517.1 JAS01822.1 KQ434823 KZC07265.1 GBGD01001590 JAC87299.1 GFDF01008358 JAV05726.1 GBBM01001216 JAC34202.1 JO843767 AEO35384.1 GBBL01001202 JAC26118.1 GBBK01002575 GBBK01002574 JAC21907.1 GFTR01006017 JAW10409.1 ACPB03021270 GFDF01008357 JAV05727.1 GEFH01003931 JAP64650.1 GFDF01008356 JAV05728.1 GFAC01003319 JAT95869.1 GFPF01012725 MAA23871.1 GEDV01007277 JAP81280.1 GACK01004745 JAA60289.1 NNAY01001596 OXU23477.1 GGLE01006765 MBY10891.1 KQ982268 KYQ58497.1 GEDC01029786 GEDC01000881 JAS07512.1 JAS36417.1 GADI01001791 JAA72017.1 GBYB01014295 JAG84062.1 GBRD01009955 GBRD01009954 JAG55870.1 GBHO01000275 GBHO01000274 JAG43329.1 JAG43330.1 GDHC01005109 GDHC01004757 JAQ13520.1 JAQ13872.1 KK114270 KFM62051.1 IAAA01036835 LAA09358.1 ABJB010152727 ABJB010394996 ABJB010473483 ABJB010762284 ABJB011053478 ABJB011062920 DS640098 EEC01796.1 GL732536 EFX83683.1 GBZX01001383 JAG91357.1 GFAH01000617 JAV47772.1 GDIP01110841 JAL92873.1 GFXV01000795 MBW12600.1 GEFM01003103 JAP72693.1 GDIQ01268204 GDIQ01252900 GDIQ01251644 GDIQ01227492 GDIQ01226344 GDIQ01224053 GDIQ01208959 GDIQ01208958 GDIQ01191318 GDIQ01168419 GDIQ01145185 GDIP01046712 GDIQ01065410 JAK83306.1 JAM57003.1 ABLF02040458 GDIP01112196 JAL91518.1 GDIP01220438 JAJ02964.1 GDIP01072548 JAM31167.1 GDIQ01179712 JAK72013.1 GDIP01072547 JAM31168.1 GDIQ01114441 JAL37285.1 GDIQ01113185 JAL38541.1 GDIQ01241846 JAK09879.1 GDIQ01179711 JAK72014.1 AJWK01033045 AJWK01033046 LBMM01010821 KMQ87207.1 GDIP01136682 JAL67032.1 APGK01037935 KB740949 ENN77191.1 GDIQ01216602 JAK35123.1 GDIQ01207664 JAK44061.1 GDIP01196937 JAJ26465.1 GDIP01254470 JAI68931.1
PCG76856.1 ODYU01008012 SOQ51263.1 MG846893 AXY94745.1 EU366906 ABY66383.1 KC481238 AGK45314.2 GAIX01003242 JAA89318.1 AGBW02009105 OWR51667.1 AK402451 BAM19073.1 AK401090 KQ459583 BAM17712.1 KPI98712.1 KU664392 ANS59102.1 GL435030 EFN74172.1 GL888137 EGI66659.1 ADTU01022136 ADTU01022137 KK107167 QOIP01000010 EZA56277.1 RLU17458.1 GL452895 EFN76608.1 GL762576 EFZ21079.1 KK852820 KDR15558.1 GECZ01025404 GECZ01012564 JAS44365.1 JAS57205.1 AK417653 BAN20868.1 KQ981891 KYN33828.1 KQ979425 KYN21607.1 KQ976394 KYM93358.1 KQ971342 EFA03968.2 JR039259 JR039260 AEY58606.1 AEY58607.1 GDJX01006590 GDJX01003127 JAT61346.1 JAT64809.1 KQ977858 KYM99329.1 GECU01011772 JAS95934.1 GEBQ01024898 JAT15079.1 NEVH01020961 PNF20334.1 KZ288349 PBC27386.1 GBYB01014298 JAG84065.1 GEZM01095793 GEZM01095792 JAV55346.1 GECZ01031842 GECZ01020215 JAS37927.1 JAS49554.1 HE962192 CCJ09440.1 GALX01004263 JAB64203.1 KF986613 AHK26791.1 LJIG01022646 KRT79463.1 BT126582 AEE61546.1 KB631923 ERL87143.1 KQ414612 KOC69173.1 KQ436240 KOX67369.1 GBBI01000195 GEMB01001334 JAC18517.1 JAS01822.1 KQ434823 KZC07265.1 GBGD01001590 JAC87299.1 GFDF01008358 JAV05726.1 GBBM01001216 JAC34202.1 JO843767 AEO35384.1 GBBL01001202 JAC26118.1 GBBK01002575 GBBK01002574 JAC21907.1 GFTR01006017 JAW10409.1 ACPB03021270 GFDF01008357 JAV05727.1 GEFH01003931 JAP64650.1 GFDF01008356 JAV05728.1 GFAC01003319 JAT95869.1 GFPF01012725 MAA23871.1 GEDV01007277 JAP81280.1 GACK01004745 JAA60289.1 NNAY01001596 OXU23477.1 GGLE01006765 MBY10891.1 KQ982268 KYQ58497.1 GEDC01029786 GEDC01000881 JAS07512.1 JAS36417.1 GADI01001791 JAA72017.1 GBYB01014295 JAG84062.1 GBRD01009955 GBRD01009954 JAG55870.1 GBHO01000275 GBHO01000274 JAG43329.1 JAG43330.1 GDHC01005109 GDHC01004757 JAQ13520.1 JAQ13872.1 KK114270 KFM62051.1 IAAA01036835 LAA09358.1 ABJB010152727 ABJB010394996 ABJB010473483 ABJB010762284 ABJB011053478 ABJB011062920 DS640098 EEC01796.1 GL732536 EFX83683.1 GBZX01001383 JAG91357.1 GFAH01000617 JAV47772.1 GDIP01110841 JAL92873.1 GFXV01000795 MBW12600.1 GEFM01003103 JAP72693.1 GDIQ01268204 GDIQ01252900 GDIQ01251644 GDIQ01227492 GDIQ01226344 GDIQ01224053 GDIQ01208959 GDIQ01208958 GDIQ01191318 GDIQ01168419 GDIQ01145185 GDIP01046712 GDIQ01065410 JAK83306.1 JAM57003.1 ABLF02040458 GDIP01112196 JAL91518.1 GDIP01220438 JAJ02964.1 GDIP01072548 JAM31167.1 GDIQ01179712 JAK72013.1 GDIP01072547 JAM31168.1 GDIQ01114441 JAL37285.1 GDIQ01113185 JAL38541.1 GDIQ01241846 JAK09879.1 GDIQ01179711 JAK72014.1 AJWK01033045 AJWK01033046 LBMM01010821 KMQ87207.1 GDIP01136682 JAL67032.1 APGK01037935 KB740949 ENN77191.1 GDIQ01216602 JAK35123.1 GDIQ01207664 JAK44061.1 GDIP01196937 JAJ26465.1 GDIP01254470 JAI68931.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000000311
UP000007755
+ More
UP000005205 UP000192223 UP000053097 UP000279307 UP000008237 UP000027135 UP000078541 UP000078492 UP000078540 UP000007266 UP000005203 UP000078542 UP000235965 UP000242457 UP000002358 UP000030742 UP000053825 UP000053105 UP000076502 UP000015103 UP000215335 UP000075809 UP000054359 UP000001555 UP000000305 UP000007819 UP000092461 UP000036403 UP000019118
UP000005205 UP000192223 UP000053097 UP000279307 UP000008237 UP000027135 UP000078541 UP000078492 UP000078540 UP000007266 UP000005203 UP000078542 UP000235965 UP000242457 UP000002358 UP000030742 UP000053825 UP000053105 UP000076502 UP000015103 UP000215335 UP000075809 UP000054359 UP000001555 UP000000305 UP000007819 UP000092461 UP000036403 UP000019118
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
Q1HPW2
Q285R3
A0A2A4JYK4
A0A2H1WDX4
A0A3G1T185
B0FYV2
+ More
N0ACL6 S4PHK3 A0A212FD75 I4DMD3 I4DIH2 A0A1B1LYS1 E1ZX82 F4WGJ4 A0A158NP28 A0A1W4XVZ8 A0A026WJQ4 E2C603 E9IEB5 A0A067R0Y2 A0A1B6F2F1 R4WDG0 A0A195F028 A0A151J9B0 A0A195BZM2 D6WKA5 A0A088AJJ6 V9ID91 A0A1D1Z3B1 A0A151IEQ7 A0A1B6J9R8 A0A1B6KUF3 A0A2J7PVH2 A0A2A3E853 A0A0C9QHZ8 A0A1Y1K7F5 A0A1B6EJ32 K4QDH4 V5GVE3 W8GNB0 A0A0T6AXY1 J3JTM5 K7J879 U4U468 A0A0L7REA5 A0A0M8ZQ94 A0A023FAM3 A0A154P5P5 A0A069DYY3 A0A1L8DH04 A0A023GJT3 G3MPG6 A0A023FZF6 A0A023FJS7 A0A224XR04 T1HS20 A0A1L8DH64 A0A131XEX0 A0A1L8DGZ6 A0A1E1X9E5 A0A224Z8H7 A0A131YS33 L7MAX7 A0A232EYW5 A0A2R5LN07 A0A151XDR0 A0A1B6C1X4 A0A0K8RLN8 A0A0C9RYG0 A0A0K8SSR9 A0A0A9ZJ75 A0A146M4N1 A0A087TAB2 A0A2L2YMJ4 B7P5C4 E9GA59 A0A0C9SCE0 A0A1W7R9F4 A0A0P5UMI5 A0A2H8TEY3 A0A131XZM9 A0A0N8BKN0 J9JQC9 A0A0P5UJ14 A0A0P4Z8M2 A0A0P5YFG9 A0A0P5KTM7 A0A0P5XGW5 A0A0P5QGP2 A0A0P5QRW3 A0A0P5GIX8 A0A0P5LJ14 A0A1B0CWZ5 A0A0J7KA78 A0A0P5SQT1 N6U958 A0A0P5I9K1 A0A0P5J364 A0A0N7ZZM5 A0A0P4WN89
N0ACL6 S4PHK3 A0A212FD75 I4DMD3 I4DIH2 A0A1B1LYS1 E1ZX82 F4WGJ4 A0A158NP28 A0A1W4XVZ8 A0A026WJQ4 E2C603 E9IEB5 A0A067R0Y2 A0A1B6F2F1 R4WDG0 A0A195F028 A0A151J9B0 A0A195BZM2 D6WKA5 A0A088AJJ6 V9ID91 A0A1D1Z3B1 A0A151IEQ7 A0A1B6J9R8 A0A1B6KUF3 A0A2J7PVH2 A0A2A3E853 A0A0C9QHZ8 A0A1Y1K7F5 A0A1B6EJ32 K4QDH4 V5GVE3 W8GNB0 A0A0T6AXY1 J3JTM5 K7J879 U4U468 A0A0L7REA5 A0A0M8ZQ94 A0A023FAM3 A0A154P5P5 A0A069DYY3 A0A1L8DH04 A0A023GJT3 G3MPG6 A0A023FZF6 A0A023FJS7 A0A224XR04 T1HS20 A0A1L8DH64 A0A131XEX0 A0A1L8DGZ6 A0A1E1X9E5 A0A224Z8H7 A0A131YS33 L7MAX7 A0A232EYW5 A0A2R5LN07 A0A151XDR0 A0A1B6C1X4 A0A0K8RLN8 A0A0C9RYG0 A0A0K8SSR9 A0A0A9ZJ75 A0A146M4N1 A0A087TAB2 A0A2L2YMJ4 B7P5C4 E9GA59 A0A0C9SCE0 A0A1W7R9F4 A0A0P5UMI5 A0A2H8TEY3 A0A131XZM9 A0A0N8BKN0 J9JQC9 A0A0P5UJ14 A0A0P4Z8M2 A0A0P5YFG9 A0A0P5KTM7 A0A0P5XGW5 A0A0P5QGP2 A0A0P5QRW3 A0A0P5GIX8 A0A0P5LJ14 A0A1B0CWZ5 A0A0J7KA78 A0A0P5SQT1 N6U958 A0A0P5I9K1 A0A0P5J364 A0A0N7ZZM5 A0A0P4WN89
PDB
3EIQ
E-value=9.70034e-165,
Score=1489
Ontologies
GO
Topology
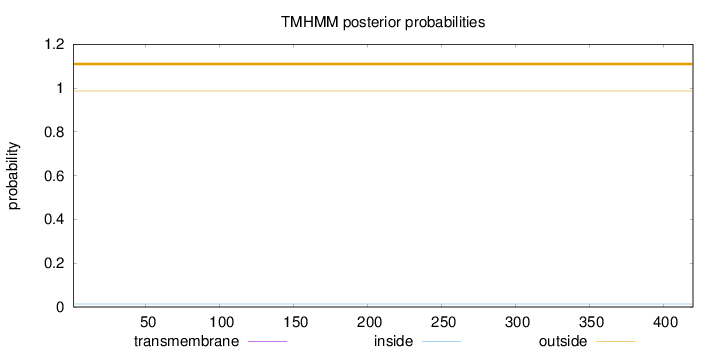
Length:
420
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00127
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01328
outside
1 - 420
Population Genetic Test Statistics
Pi
221.755508
Theta
208.818635
Tajima's D
0.198898
CLR
0.067164
CSRT
0.419829008549573
Interpretation
Uncertain
