Gene
KWMTBOMO02080
Pre Gene Modal
BGIBMGA002980
Annotation
PREDICTED:_uncharacterized_protein_LOC101741263_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.064 PlasmaMembrane Reliability : 1.913
Sequence
CDS
ATGCGTGGTAGTCGTAGGGTAGTGATTTTTTGCTTATTGACTACGCTGCTACCTACAACGCTGATAGTGATCCCTTTATATTTAAGAAATATTAAATATGCTGATGTTATGTATCGCATATCAGATTCTGATGTAATACAAATACACAAAGGACAATCTTCAGTTTTCTGTGAGAAACACACACTACAAATGAATACTTCTTTTAATGCTTTTCAAATGAGTGGCATACCGGAACTTTCAGAGAAAAGAAAGCACATTCGACTGAAAAAATCAATGACTCTGCCAGATGACACTTTGGAATACTGGGGTTTTTATCTGTTAAAAGGCGCCACAGTAGAACTCAAGGCCTGTTCAAGATATGATGGCTCAAAAATCTTAGTTGTAAAAGGTGACAGAATACTGAATACATGTGGTATTTTAGACAGACGAAAATTTAAGAAAGAACCTTCATTAGCCGAAAATCAAGGAACTGTACTGGTAACATTACAAAATCCTAATAACAAAAAGGAGAGCGAAATTTATGCCAGAAGTACTCACCGTGAAAAGGCCAAGGATCCGTTGGACGAAGACATACAAACAAAACCTAAATTTGAAAATATTTACTTGAAATCACAATTTACATCAGGCACAAATAAAAAGCAGCAAAAAAGAGATACACGCACCAAATTGCATAATCCTAATACTATTCTTGATACAGGAGTACATCACGGCGGGAATGCGTTTAACAGTACTGTCAAGCCAGACAATAGTGCTGTATCCAGTTTCGAGATAAATTTATACGATTGCTACCAAGATAATATATTGATGAATGAGTATTTTCCCTTTTCAATTCAGTGTAATAGTACAAACTATTTGGAAGAGACTACAATTTTAAAAGTTATTCATGAAGTGTTAGTGGATGGATATTACTATTACATGTTTTATAGTGATAACGATATTGTATTTAACGACATTCATGCAATTTTTGACATTTATAAGCCTACATTCCAATATGTCAATAACTCTAGTTCCAAAATGTGCATCAATAAAACAAGATGTGAATTTGATATAGGGTTTTTTAGTGACGAACTTGTTATTGTAGAAGTGCCAACAAAAGGCGGATTGCGGAACGATGAGGACGATACGTCTGTTCTAATTTCCGTTTGTCTTCCCAGAAAATCCGTTTACATATTTTTCCCCATTGCTGTTCTTCTTCTGATACTACTTTTTGCATTTTTATGA
Protein
MRGSRRVVIFCLLTTLLPTTLIVIPLYLRNIKYADVMYRISDSDVIQIHKGQSSVFCEKHTLQMNTSFNAFQMSGIPELSEKRKHIRLKKSMTLPDDTLEYWGFYLLKGATVELKACSRYDGSKILVVKGDRILNTCGILDRRKFKKEPSLAENQGTVLVTLQNPNNKKESEIYARSTHREKAKDPLDEDIQTKPKFENIYLKSQFTSGTNKKQQKRDTRTKLHNPNTILDTGVHHGGNAFNSTVKPDNSAVSSFEINLYDCYQDNILMNEYFPFSIQCNSTNYLEETTILKVIHEVLVDGYYYYMFYSDNDIVFNDIHAIFDIYKPTFQYVNNSSSKMCINKTRCEFDIGFFSDELVIVEVPTKGGLRNDEDDTSVLISVCLPRKSVYIFFPIAVLLLILLFAFL
Summary
Uniprot
H9J0E4
A0A0L7LKY6
A0A1E1W3B8
A0A2A4JZN6
A0A2A4JXW4
A0A194Q088
+ More
A0A2H1WEI3 A0A194QLN8 S4PYD0 A0A1L8D9V9 A0A1L8D9G4 D6WKA4 A0A1A9W868 T1GJ29 A0A1A9UIV6 A0A1A9XWR3 A0A336M1Q3 B4KFA3 A0A0M4EDR5 A0A0Q9XG82 A0A0Q9W821 A0A0Q9WI78 A0A0Q9XHC8 A0A1B0BP46 B4LUY3 V5GQU2 V5I8K6 B4MUK2 A0A0A9XYA8 A0A1B6D7I7 A0A1B6CL27 A0A3B0JXN1 A0A0K8SLU4 A0A224XNV8 B4GJ77 Q29JS7 A0A0R3NVR6 A0A1B6H3A0 A0A0R3P0I0 A0A0T6AWD0 A0A1B6IIV3 A0A1B6HWY8 A0A1W4XKU3 A0A1Y1JUB4 A0A1W4XVV7 A0A0N8NZF9 B3MUQ0 A0A0P8ZHP1 A0A1B0CK22 B4JZE3 A0A2R7WL89
A0A2H1WEI3 A0A194QLN8 S4PYD0 A0A1L8D9V9 A0A1L8D9G4 D6WKA4 A0A1A9W868 T1GJ29 A0A1A9UIV6 A0A1A9XWR3 A0A336M1Q3 B4KFA3 A0A0M4EDR5 A0A0Q9XG82 A0A0Q9W821 A0A0Q9WI78 A0A0Q9XHC8 A0A1B0BP46 B4LUY3 V5GQU2 V5I8K6 B4MUK2 A0A0A9XYA8 A0A1B6D7I7 A0A1B6CL27 A0A3B0JXN1 A0A0K8SLU4 A0A224XNV8 B4GJ77 Q29JS7 A0A0R3NVR6 A0A1B6H3A0 A0A0R3P0I0 A0A0T6AWD0 A0A1B6IIV3 A0A1B6HWY8 A0A1W4XKU3 A0A1Y1JUB4 A0A1W4XVV7 A0A0N8NZF9 B3MUQ0 A0A0P8ZHP1 A0A1B0CK22 B4JZE3 A0A2R7WL89
Pubmed
EMBL
BABH01010502
JTDY01000719
KOB76102.1
GDQN01009635
JAT81419.1
NWSH01000373
+ More
PCG76862.1 PCG76861.1 KQ459583 KPI98713.1 ODYU01008012 SOQ51262.1 KQ461196 KPJ06473.1 GAIX01003503 JAA89057.1 GFDF01010974 JAV03110.1 GFDF01010992 JAV03092.1 KQ971342 EFA03967.1 CAQQ02058401 UFQT01000281 SSX22753.1 CH933807 EDW12003.1 KRG03033.1 CP012523 ALC40035.1 KRG03031.1 CH940649 KRF80979.1 KRF80980.1 KRG03032.1 JXJN01017808 EDW63232.1 GALX01004519 JAB63947.1 GALX01004520 JAB63946.1 CH963857 EDW76197.2 GBHO01019238 GDHC01011653 GDHC01003034 JAG24366.1 JAQ06976.1 JAQ15595.1 GEDC01015763 JAS21535.1 GEDC01023148 JAS14150.1 OUUW01000010 SPP85823.1 GBRD01011596 GBRD01011591 GBRD01011514 GBRD01004938 GBRD01004937 JAG54228.1 GFTR01006174 JAW10252.1 CH479184 EDW37391.1 CH379062 EAL32884.2 KRT05036.1 GECZ01000607 JAS69162.1 KRT05037.1 LJIG01022646 KRT79459.1 GECU01020852 JAS86854.1 GECU01028517 JAS79189.1 GEZM01100402 JAV52892.1 CH902624 KPU74212.1 EDV32965.2 KPU74211.1 AJWK01015648 CH916379 EDV94065.1 KK854931 PTY19760.1
PCG76862.1 PCG76861.1 KQ459583 KPI98713.1 ODYU01008012 SOQ51262.1 KQ461196 KPJ06473.1 GAIX01003503 JAA89057.1 GFDF01010974 JAV03110.1 GFDF01010992 JAV03092.1 KQ971342 EFA03967.1 CAQQ02058401 UFQT01000281 SSX22753.1 CH933807 EDW12003.1 KRG03033.1 CP012523 ALC40035.1 KRG03031.1 CH940649 KRF80979.1 KRF80980.1 KRG03032.1 JXJN01017808 EDW63232.1 GALX01004519 JAB63947.1 GALX01004520 JAB63946.1 CH963857 EDW76197.2 GBHO01019238 GDHC01011653 GDHC01003034 JAG24366.1 JAQ06976.1 JAQ15595.1 GEDC01015763 JAS21535.1 GEDC01023148 JAS14150.1 OUUW01000010 SPP85823.1 GBRD01011596 GBRD01011591 GBRD01011514 GBRD01004938 GBRD01004937 JAG54228.1 GFTR01006174 JAW10252.1 CH479184 EDW37391.1 CH379062 EAL32884.2 KRT05036.1 GECZ01000607 JAS69162.1 KRT05037.1 LJIG01022646 KRT79459.1 GECU01020852 JAS86854.1 GECU01028517 JAS79189.1 GEZM01100402 JAV52892.1 CH902624 KPU74212.1 EDV32965.2 KPU74211.1 AJWK01015648 CH916379 EDV94065.1 KK854931 PTY19760.1
Proteomes
PRIDE
SUPFAM
SSF54695
SSF54695
ProteinModelPortal
H9J0E4
A0A0L7LKY6
A0A1E1W3B8
A0A2A4JZN6
A0A2A4JXW4
A0A194Q088
+ More
A0A2H1WEI3 A0A194QLN8 S4PYD0 A0A1L8D9V9 A0A1L8D9G4 D6WKA4 A0A1A9W868 T1GJ29 A0A1A9UIV6 A0A1A9XWR3 A0A336M1Q3 B4KFA3 A0A0M4EDR5 A0A0Q9XG82 A0A0Q9W821 A0A0Q9WI78 A0A0Q9XHC8 A0A1B0BP46 B4LUY3 V5GQU2 V5I8K6 B4MUK2 A0A0A9XYA8 A0A1B6D7I7 A0A1B6CL27 A0A3B0JXN1 A0A0K8SLU4 A0A224XNV8 B4GJ77 Q29JS7 A0A0R3NVR6 A0A1B6H3A0 A0A0R3P0I0 A0A0T6AWD0 A0A1B6IIV3 A0A1B6HWY8 A0A1W4XKU3 A0A1Y1JUB4 A0A1W4XVV7 A0A0N8NZF9 B3MUQ0 A0A0P8ZHP1 A0A1B0CK22 B4JZE3 A0A2R7WL89
A0A2H1WEI3 A0A194QLN8 S4PYD0 A0A1L8D9V9 A0A1L8D9G4 D6WKA4 A0A1A9W868 T1GJ29 A0A1A9UIV6 A0A1A9XWR3 A0A336M1Q3 B4KFA3 A0A0M4EDR5 A0A0Q9XG82 A0A0Q9W821 A0A0Q9WI78 A0A0Q9XHC8 A0A1B0BP46 B4LUY3 V5GQU2 V5I8K6 B4MUK2 A0A0A9XYA8 A0A1B6D7I7 A0A1B6CL27 A0A3B0JXN1 A0A0K8SLU4 A0A224XNV8 B4GJ77 Q29JS7 A0A0R3NVR6 A0A1B6H3A0 A0A0R3P0I0 A0A0T6AWD0 A0A1B6IIV3 A0A1B6HWY8 A0A1W4XKU3 A0A1Y1JUB4 A0A1W4XVV7 A0A0N8NZF9 B3MUQ0 A0A0P8ZHP1 A0A1B0CK22 B4JZE3 A0A2R7WL89
Ontologies
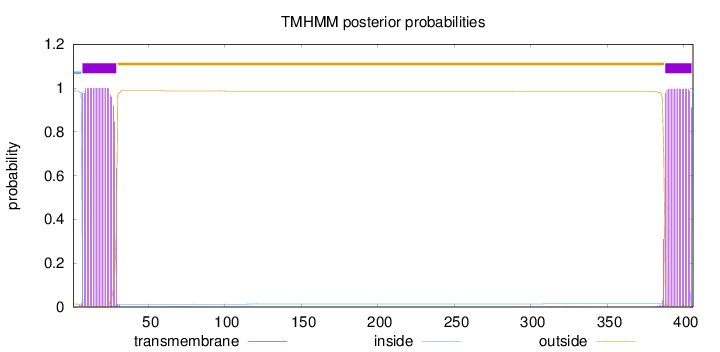
Topology
Length:
406
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.54387
Exp number, first 60 AAs:
22.32333
Total prob of N-in:
0.98684
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 387
TMhelix
388 - 405
inside
406 - 406
Population Genetic Test Statistics
Pi
210.0453
Theta
189.291925
Tajima's D
0.306847
CLR
0.658987
CSRT
0.461426928653567
Interpretation
Uncertain
