Gene
KWMTBOMO02076 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002979
Annotation
PREDICTED:_carbonyl_reductase_[NADPH]_1-like_[Amyelois_transitella]
Full name
Transmembrane 9 superfamily member
Location in the cell
Cytoplasmic Reliability : 2.965
Sequence
CDS
ATGGAAACCAAAGTTGCAGTAGTAACTGGAAGCAATAAAGGAATAGGCTTTGAAATCGTTAAAGGATTGTGTCAGAAGTTTAATGGTGTAGTGTACCTCACAGCTCGAAATGAAGAAAGAGGGCGGCAAGCTATTAGGAAGCTACGAGAGTTCGAGTTAAATCCTTTATTTCATCTGCTGGATGTAACAGATGAAGAAAGTATAAAAGAGTTCGCTTCCCATATTTCAAAACACCATTCGGGTATTGATGTATTCGTAAACAATGCCGGCATTCTGGACTTTGATAAGTCTGTACCTTCCTATGAAGACGCTAAGAAATTAATCGACACGAATTTCATGAGCTTATTAACCATATCAAAAATATTATACCCTCTACTGAAAAACGATGCACGAATTATAAATCTAAGTAGTGATTGGGGTTTGTTATCGAATATCAGGAAACAAGTGTGGTTAGATACGCTCGTAAAGGATGATCTTACAGTGGAAGAAATATTGCAATTTGTTAACGATTTCTTAACTGCTGTGAAAGGTGGTAAAAATACATTTGTATCGTTTGCGGGTCAGCATGGCGATTACAAAGTTTCAAAAGTGGCATTGTCTGCGTTAACCTTTGTGCAACAGAGACAATTTACTGAACAAGGGAAAGATATTTCAGTTAATTGTGTTCACCCTGGGTTCATTAAAACTGATATGACAAAAGGAATGGGTGATTTTGAACCTGAAAGAGGTGCACGAGCTCCACTTTATTTGGCCCTAGAAGCACCTCAGTCACAGAAAGGCACATTTGTGTGGCATGATTGTCATCAAGTAAATTGGGACGATTGA
Protein
METKVAVVTGSNKGIGFEIVKGLCQKFNGVVYLTARNEERGRQAIRKLREFELNPLFHLLDVTDEESIKEFASHISKHHSGIDVFVNNAGILDFDKSVPSYEDAKKLIDTNFMSLLTISKILYPLLKNDARIINLSSDWGLLSNIRKQVWLDTLVKDDLTVEEILQFVNDFLTAVKGGKNTFVSFAGQHGDYKVSKVALSALTFVQQRQFTEQGKDISVNCVHPGFIKTDMTKGMGDFEPERGARAPLYLALEAPQSQKGTFVWHDCHQVNWDD
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Belongs to the nonaspanin (TM9SF) (TC 9.A.2) family.
Belongs to the nonaspanin (TM9SF) (TC 9.A.2) family.
Uniprot
H9J0E3
A0A2H1WXI4
S4PFV4
A0A194QM92
A0A194PJW2
A0A212FD48
+ More
A0A0L7LKQ9 A0A0N0PAU9 A0A194QFC8 A0A2H1WY85 A0A0L7L0Z3 A0A2W1BHX9 A0A2A4IZP8 A0A2A4IS40 A0A2A4JMV1 S4PV51 H9J991 D2SNQ5 A0A2W1BQ78 A9NX88 A0A2A4JNM1 A0A2W1BQF4 A0A194PYN6 A0A194RBM8 A0A212FD24 A0A212FD16 A0A194R7T9 A0A194Q0C3 A0A2H1VKT8 A0A194PZI3 A0A194PYP0 A0A194REE5 A0A0L7L987 A0A194QI05 A0A194Q4V7 V5GN34 A0A1W4X692 A0A194PVS9 A0A0N1IER6 L7M5Y8 A0A1Y1NF00 A0A2A4IVK8 A0A2H1VYG0 A0A0K8REZ9 H9JV01 A0A1B6FR44 A0A1B6CF00 A0A131XBH7 D6WUE0 A0A1B6ITF8 J3JTF1 A0A0N1I826 A0A023FD31 A0A1B6M0H7 A0A224YPN9 A0A224YJ66 A0A0K8R6T0 A0A0L7KMS8 L7M4D0 A0A131XVV1 A0A023GHB1 A0A1W4WY33 A0A1B6CFB5 A0A131Z4Y0 G3MQJ3 A0A023GKM9 A0A224YLL4 A0A023GK08 A0A1Z5KVU9 A0A2H1VYG7 A0A1B6IYN9 A0A1B6C7K7 A0A023GHC5 A0A2J7Q296 A0A131XF63 A0A293L6R9 A0A224YG15 A0A087UKN1 A0A1Y1KDA4 A0A1B6M714 A0A2R7VQP4 A0A023F803 A0A194R6L7 A0A2S2QWN2 A0A1B6EX40 V5HFZ7 A0A1E1XAN7 A0A2R5LLH9 A0A1B6KVH8 G3MJH5 A0A2H1VPT5 T1L244 J3JU59 R4FNV0 A0A023FKP1 A0A023FHX5 A0A224XVD1 A0A0P5DYT1 A0A0P5ZIU5 A0A0P4YT29
A0A0L7LKQ9 A0A0N0PAU9 A0A194QFC8 A0A2H1WY85 A0A0L7L0Z3 A0A2W1BHX9 A0A2A4IZP8 A0A2A4IS40 A0A2A4JMV1 S4PV51 H9J991 D2SNQ5 A0A2W1BQ78 A9NX88 A0A2A4JNM1 A0A2W1BQF4 A0A194PYN6 A0A194RBM8 A0A212FD24 A0A212FD16 A0A194R7T9 A0A194Q0C3 A0A2H1VKT8 A0A194PZI3 A0A194PYP0 A0A194REE5 A0A0L7L987 A0A194QI05 A0A194Q4V7 V5GN34 A0A1W4X692 A0A194PVS9 A0A0N1IER6 L7M5Y8 A0A1Y1NF00 A0A2A4IVK8 A0A2H1VYG0 A0A0K8REZ9 H9JV01 A0A1B6FR44 A0A1B6CF00 A0A131XBH7 D6WUE0 A0A1B6ITF8 J3JTF1 A0A0N1I826 A0A023FD31 A0A1B6M0H7 A0A224YPN9 A0A224YJ66 A0A0K8R6T0 A0A0L7KMS8 L7M4D0 A0A131XVV1 A0A023GHB1 A0A1W4WY33 A0A1B6CFB5 A0A131Z4Y0 G3MQJ3 A0A023GKM9 A0A224YLL4 A0A023GK08 A0A1Z5KVU9 A0A2H1VYG7 A0A1B6IYN9 A0A1B6C7K7 A0A023GHC5 A0A2J7Q296 A0A131XF63 A0A293L6R9 A0A224YG15 A0A087UKN1 A0A1Y1KDA4 A0A1B6M714 A0A2R7VQP4 A0A023F803 A0A194R6L7 A0A2S2QWN2 A0A1B6EX40 V5HFZ7 A0A1E1XAN7 A0A2R5LLH9 A0A1B6KVH8 G3MJH5 A0A2H1VPT5 T1L244 J3JU59 R4FNV0 A0A023FKP1 A0A023FHX5 A0A224XVD1 A0A0P5DYT1 A0A0P5ZIU5 A0A0P4YT29
Pubmed
EMBL
BABH01010518
ODYU01011824
SOQ57785.1
GAIX01003872
JAA88688.1
KQ461196
+ More
KPJ06479.1 KQ459601 KPI93721.1 AGBW02009105 OWR51675.1 JTDY01000815 KOB75776.1 KQ461157 KPJ08086.1 KQ459053 KPJ04248.1 ODYU01011980 SOQ58033.1 JTDY01003824 KOB68949.1 KZ150152 PZC72817.1 NWSH01004922 PCG64583.1 NWSH01008342 PCG62595.1 NWSH01000963 PCG73377.1 GAIX01009148 JAA83412.1 BABH01004306 EZ407185 ACX53746.1 KZ149934 PZC77229.1 EF085953 ABK25249.1 PCG73376.1 PZC77228.1 KQ459584 KPI98447.1 KQ460644 KPJ13306.1 AGBW02009113 OWR51646.1 OWR51645.1 KPJ13305.1 KPI98444.1 ODYU01003092 SOQ41411.1 KPI98448.1 KPI98446.1 KQ460325 KPJ15839.1 JTDY01002114 KOB72078.1 KQ459193 KPJ03086.1 KPI98445.1 GALX01002952 JAB65514.1 KQ459597 KPI95235.1 KQ460466 KPJ14619.1 GACK01005752 JAA59282.1 GEZM01008630 JAV94726.1 NWSH01006830 PCG63222.1 ODYU01005228 SOQ45880.1 GADI01004071 JAA69737.1 BABH01037460 GECZ01017087 GECZ01016387 JAS52682.1 JAS53382.1 GEDC01025309 JAS11989.1 GEFH01004098 JAP64483.1 KQ971352 EFA07299.1 GECU01027825 GECU01017498 JAS79881.1 JAS90208.1 BT126507 AEE61471.1 KPJ14621.1 GBBK01005242 JAC19240.1 GEBQ01010531 JAT29446.1 GFPF01006553 MAA17699.1 GFPF01006552 MAA17698.1 GADI01006938 JAA66870.1 JTDY01008746 KOB64420.1 GACK01007031 JAA58003.1 GEFM01004999 JAP70797.1 GBBM01002052 JAC33366.1 GEDC01025189 JAS12109.1 GEDV01003386 JAP85171.1 JO844144 AEO35761.1 GBBM01002053 JAC33365.1 GFPF01005493 MAA16639.1 GBBM01002050 JAC33368.1 GFJQ02007744 JAV99225.1 SOQ45879.1 GECU01015736 JAS91970.1 GEDC01027847 GEDC01011823 JAS09451.1 JAS25475.1 GBBM01002051 JAC33367.1 NEVH01019373 PNF22703.1 GEFH01002778 JAP65803.1 GFWV01003337 MAA28067.1 GFPF01005492 MAA16638.1 KK120274 KFM77920.1 GEZM01088470 JAV58180.1 GEBQ01008269 JAT31708.1 KK854031 PTY09833.1 GBBI01001358 JAC17354.1 KPJ13307.1 GGMS01012953 MBY82156.1 GECZ01027302 JAS42467.1 GANP01008039 JAB76429.1 GFAC01003037 JAT96151.1 GGLE01006223 MBY10349.1 GEBQ01024723 JAT15254.1 JO842026 AEO33643.1 ODYU01003722 SOQ42855.1 CAEY01000925 BT126771 AEE61733.1 ACPB03015037 GAHY01001144 JAA76366.1 GBBK01003249 JAC21233.1 GBBK01003420 JAC21062.1 GFTR01004535 JAW11891.1 GDIP01149008 JAJ74394.1 GDIP01044472 JAM59243.1 GDIP01223444 JAI99957.1
KPJ06479.1 KQ459601 KPI93721.1 AGBW02009105 OWR51675.1 JTDY01000815 KOB75776.1 KQ461157 KPJ08086.1 KQ459053 KPJ04248.1 ODYU01011980 SOQ58033.1 JTDY01003824 KOB68949.1 KZ150152 PZC72817.1 NWSH01004922 PCG64583.1 NWSH01008342 PCG62595.1 NWSH01000963 PCG73377.1 GAIX01009148 JAA83412.1 BABH01004306 EZ407185 ACX53746.1 KZ149934 PZC77229.1 EF085953 ABK25249.1 PCG73376.1 PZC77228.1 KQ459584 KPI98447.1 KQ460644 KPJ13306.1 AGBW02009113 OWR51646.1 OWR51645.1 KPJ13305.1 KPI98444.1 ODYU01003092 SOQ41411.1 KPI98448.1 KPI98446.1 KQ460325 KPJ15839.1 JTDY01002114 KOB72078.1 KQ459193 KPJ03086.1 KPI98445.1 GALX01002952 JAB65514.1 KQ459597 KPI95235.1 KQ460466 KPJ14619.1 GACK01005752 JAA59282.1 GEZM01008630 JAV94726.1 NWSH01006830 PCG63222.1 ODYU01005228 SOQ45880.1 GADI01004071 JAA69737.1 BABH01037460 GECZ01017087 GECZ01016387 JAS52682.1 JAS53382.1 GEDC01025309 JAS11989.1 GEFH01004098 JAP64483.1 KQ971352 EFA07299.1 GECU01027825 GECU01017498 JAS79881.1 JAS90208.1 BT126507 AEE61471.1 KPJ14621.1 GBBK01005242 JAC19240.1 GEBQ01010531 JAT29446.1 GFPF01006553 MAA17699.1 GFPF01006552 MAA17698.1 GADI01006938 JAA66870.1 JTDY01008746 KOB64420.1 GACK01007031 JAA58003.1 GEFM01004999 JAP70797.1 GBBM01002052 JAC33366.1 GEDC01025189 JAS12109.1 GEDV01003386 JAP85171.1 JO844144 AEO35761.1 GBBM01002053 JAC33365.1 GFPF01005493 MAA16639.1 GBBM01002050 JAC33368.1 GFJQ02007744 JAV99225.1 SOQ45879.1 GECU01015736 JAS91970.1 GEDC01027847 GEDC01011823 JAS09451.1 JAS25475.1 GBBM01002051 JAC33367.1 NEVH01019373 PNF22703.1 GEFH01002778 JAP65803.1 GFWV01003337 MAA28067.1 GFPF01005492 MAA16638.1 KK120274 KFM77920.1 GEZM01088470 JAV58180.1 GEBQ01008269 JAT31708.1 KK854031 PTY09833.1 GBBI01001358 JAC17354.1 KPJ13307.1 GGMS01012953 MBY82156.1 GECZ01027302 JAS42467.1 GANP01008039 JAB76429.1 GFAC01003037 JAT96151.1 GGLE01006223 MBY10349.1 GEBQ01024723 JAT15254.1 JO842026 AEO33643.1 ODYU01003722 SOQ42855.1 CAEY01000925 BT126771 AEE61733.1 ACPB03015037 GAHY01001144 JAA76366.1 GBBK01003249 JAC21233.1 GBBK01003420 JAC21062.1 GFTR01004535 JAW11891.1 GDIP01149008 JAJ74394.1 GDIP01044472 JAM59243.1 GDIP01223444 JAI99957.1
Proteomes
Interpro
SUPFAM
SSF51735
SSF51735
CDD
ProteinModelPortal
H9J0E3
A0A2H1WXI4
S4PFV4
A0A194QM92
A0A194PJW2
A0A212FD48
+ More
A0A0L7LKQ9 A0A0N0PAU9 A0A194QFC8 A0A2H1WY85 A0A0L7L0Z3 A0A2W1BHX9 A0A2A4IZP8 A0A2A4IS40 A0A2A4JMV1 S4PV51 H9J991 D2SNQ5 A0A2W1BQ78 A9NX88 A0A2A4JNM1 A0A2W1BQF4 A0A194PYN6 A0A194RBM8 A0A212FD24 A0A212FD16 A0A194R7T9 A0A194Q0C3 A0A2H1VKT8 A0A194PZI3 A0A194PYP0 A0A194REE5 A0A0L7L987 A0A194QI05 A0A194Q4V7 V5GN34 A0A1W4X692 A0A194PVS9 A0A0N1IER6 L7M5Y8 A0A1Y1NF00 A0A2A4IVK8 A0A2H1VYG0 A0A0K8REZ9 H9JV01 A0A1B6FR44 A0A1B6CF00 A0A131XBH7 D6WUE0 A0A1B6ITF8 J3JTF1 A0A0N1I826 A0A023FD31 A0A1B6M0H7 A0A224YPN9 A0A224YJ66 A0A0K8R6T0 A0A0L7KMS8 L7M4D0 A0A131XVV1 A0A023GHB1 A0A1W4WY33 A0A1B6CFB5 A0A131Z4Y0 G3MQJ3 A0A023GKM9 A0A224YLL4 A0A023GK08 A0A1Z5KVU9 A0A2H1VYG7 A0A1B6IYN9 A0A1B6C7K7 A0A023GHC5 A0A2J7Q296 A0A131XF63 A0A293L6R9 A0A224YG15 A0A087UKN1 A0A1Y1KDA4 A0A1B6M714 A0A2R7VQP4 A0A023F803 A0A194R6L7 A0A2S2QWN2 A0A1B6EX40 V5HFZ7 A0A1E1XAN7 A0A2R5LLH9 A0A1B6KVH8 G3MJH5 A0A2H1VPT5 T1L244 J3JU59 R4FNV0 A0A023FKP1 A0A023FHX5 A0A224XVD1 A0A0P5DYT1 A0A0P5ZIU5 A0A0P4YT29
A0A0L7LKQ9 A0A0N0PAU9 A0A194QFC8 A0A2H1WY85 A0A0L7L0Z3 A0A2W1BHX9 A0A2A4IZP8 A0A2A4IS40 A0A2A4JMV1 S4PV51 H9J991 D2SNQ5 A0A2W1BQ78 A9NX88 A0A2A4JNM1 A0A2W1BQF4 A0A194PYN6 A0A194RBM8 A0A212FD24 A0A212FD16 A0A194R7T9 A0A194Q0C3 A0A2H1VKT8 A0A194PZI3 A0A194PYP0 A0A194REE5 A0A0L7L987 A0A194QI05 A0A194Q4V7 V5GN34 A0A1W4X692 A0A194PVS9 A0A0N1IER6 L7M5Y8 A0A1Y1NF00 A0A2A4IVK8 A0A2H1VYG0 A0A0K8REZ9 H9JV01 A0A1B6FR44 A0A1B6CF00 A0A131XBH7 D6WUE0 A0A1B6ITF8 J3JTF1 A0A0N1I826 A0A023FD31 A0A1B6M0H7 A0A224YPN9 A0A224YJ66 A0A0K8R6T0 A0A0L7KMS8 L7M4D0 A0A131XVV1 A0A023GHB1 A0A1W4WY33 A0A1B6CFB5 A0A131Z4Y0 G3MQJ3 A0A023GKM9 A0A224YLL4 A0A023GK08 A0A1Z5KVU9 A0A2H1VYG7 A0A1B6IYN9 A0A1B6C7K7 A0A023GHC5 A0A2J7Q296 A0A131XF63 A0A293L6R9 A0A224YG15 A0A087UKN1 A0A1Y1KDA4 A0A1B6M714 A0A2R7VQP4 A0A023F803 A0A194R6L7 A0A2S2QWN2 A0A1B6EX40 V5HFZ7 A0A1E1XAN7 A0A2R5LLH9 A0A1B6KVH8 G3MJH5 A0A2H1VPT5 T1L244 J3JU59 R4FNV0 A0A023FKP1 A0A023FHX5 A0A224XVD1 A0A0P5DYT1 A0A0P5ZIU5 A0A0P4YT29
PDB
1N5D
E-value=1.97035e-40,
Score=415
Ontologies
PATHWAY
PANTHER
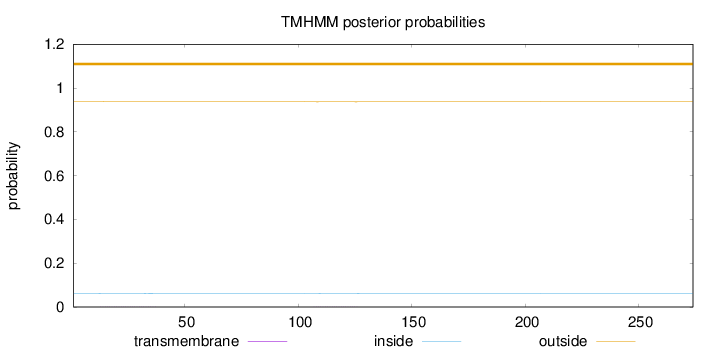
Topology
Length:
274
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03219
Exp number, first 60 AAs:
0.01995
Total prob of N-in:
0.06206
outside
1 - 274
Population Genetic Test Statistics
Pi
345.131437
Theta
237.159914
Tajima's D
1.433368
CLR
0
CSRT
0.774261286935653
Interpretation
Uncertain
