Gene
KWMTBOMO02075
Pre Gene Modal
BGIBMGA002978
Annotation
PREDICTED:_transmembrane_9_superfamily_member_4_[Papilio_xuthus]
Full name
Transmembrane 9 superfamily member
Location in the cell
PlasmaMembrane Reliability : 4.965
Sequence
CDS
ATGTTTCAAGTTTATCTTCAGTGGATTCTATTCATTACCATAATTTTAAAAAGTTATGGCTTCTATGTACCTGGTGTGGCACCGGTCGAGTTCAAAAAAGGTCAAAAAATTGAAGTAAAAGCTGTTAAGATGACCAGCATTCATACTCAATTACCATACGAATATTATTCATTGCCTTTGTGTCTCCCAAAAAATGGCACATTTGTGTACAAATCTGAAAACTTAGGAGAAGTTTTGCGCGGAGATCGTATCGTCAATACACCATATGAAGTTCATATGGCAGAGAACATTAAATGCAAACTTCTTTGCCACAGGAAAAACGTTCCTATCAATTGGAGTGTAGAAGATTCTGAAAAAGTTGCTAGTCGTATTGAACATGAATATTTTGTTCACTTATTAGTTGATAATTTACCTGTTGCAACAAAAGTTATAAATGTTGAAACTCTAGAAAGGACTATTGAGCAAGGATATCGTCTTGGATATATGTCTGAAGGAAAGGCCTACATTAATAACCATCTTAAACTGTTACTGAAGTACCATAAGCATAGCCAGGATTCTTATAGAGTAGTGGGATTTGAAGTTGAAACCCATTCAGTTTCTAAAGATGAATTAATATTTACTGATGATACTTGTCAAATACCAGTCAAACCAAAGCCACAGTTAGTTAATGAAGATACTGGTACAAAGTTGTACTTCACATACTCTGTGGAATGGGGAGAATCAGATATTAGCTGGGCGTCAAGATGGGACATTTATTTGGGCATGAAGGATGTACAGATACATTGGTTCTCTATTATTAATTCTGTTGTTGTATTATTTTTCTTGTCAGCCATATTAACAATGATAATGGTACGAACATTGAGACGAGACATAGCTCGCTATAATTCAGTTGAAGGAATTGATGACATGATGGAAGAAACAGGATGGAAATTAGTACATGGAGATGTCTTCCGACCTCCTCCTAAGAGGATGTTGTTTGCAGCTGTGGTGGGAAGTGGGATACAAGTCTTTCTGATGGCATTGATAACAATATTTATCGCGATGCTTGGTATGCTGTCGCCGGCAAGTCGAGGGGCCTTAATGACCGCCGCGATTTTCCTATACGTATTTATGGGGTTGATCGCGGGTTATTTCTCGGCGCGCCTCTATAATACAATGAAAGGCAAACAATGGAAACAAGCCGCATTCCTGACGGCAACGCTGTACCCCGCCATTGTCTTTGGCACATGTTTCTTTTTAAACTTCTTTATAATGGGCAAACATTCCAGCGGTGCTGTACCGTTTTCGACTATGATGGCCCTATTGTGTTTATGGTTTTGTATTTCATTACCGTTGGTTTATTTGGGTTATTATTTCGGATGCCGAAAACAGCCTTTCCAACACCCTGTAAGGACAAATTTCATTCCCAGGAAAATACCTGAACAAGTTTGGTATATGAATGTATTGATATGCGTTTTGATGGCAGGAATACTGCCCTTCGGAGCCGTATACATCGAGTTGTTCTTCATCTACAACGCACTTTGGGAGAACCAATTCTATTATCTTTTTGGATTTTTATTTTTAGTATTTTGCATTTTGGTTGTGTCCGTATCTCAGATATCGATATGTATGGTTTACTTCCAACTCTGCGGTGAGGACTACCACTGGTGGTGGAAAAGCTTCTTTGTATCAGGTGGATCTGCAGTGTATATATTGATTTATTCAATATTCTATTTCTTCAAGAAATTAGAGATAACAGAATTTATACCGACGTTACTATACATAGGTTATACTGGTCTCATGGTATTGACTTTTTGGCTTCTTACCGGTACCATTGGATTTTTCGCAGCATATACATTTATTAGAAAAATATACGCTGCTGTAAAAATAGATTAA
Protein
MFQVYLQWILFITIILKSYGFYVPGVAPVEFKKGQKIEVKAVKMTSIHTQLPYEYYSLPLCLPKNGTFVYKSENLGEVLRGDRIVNTPYEVHMAENIKCKLLCHRKNVPINWSVEDSEKVASRIEHEYFVHLLVDNLPVATKVINVETLERTIEQGYRLGYMSEGKAYINNHLKLLLKYHKHSQDSYRVVGFEVETHSVSKDELIFTDDTCQIPVKPKPQLVNEDTGTKLYFTYSVEWGESDISWASRWDIYLGMKDVQIHWFSIINSVVVLFFLSAILTMIMVRTLRRDIARYNSVEGIDDMMEETGWKLVHGDVFRPPPKRMLFAAVVGSGIQVFLMALITIFIAMLGMLSPASRGALMTAAIFLYVFMGLIAGYFSARLYNTMKGKQWKQAAFLTATLYPAIVFGTCFFLNFFIMGKHSSGAVPFSTMMALLCLWFCISLPLVYLGYYFGCRKQPFQHPVRTNFIPRKIPEQVWYMNVLICVLMAGILPFGAVYIELFFIYNALWENQFYYLFGFLFLVFCILVVSVSQISICMVYFQLCGEDYHWWWKSFFVSGGSAVYILIYSIFYFFKKLEITEFIPTLLYIGYTGLMVLTFWLLTGTIGFFAAYTFIRKIYAAVKID
Summary
Similarity
Belongs to the nonaspanin (TM9SF) (TC 9.A.2) family.
Feature
chain Transmembrane 9 superfamily member
Uniprot
H9J0E2
A0A2H1WXU4
A0A212FD46
A0A194QND0
A0A1E1WC95
A0A194PK74
+ More
A0A067R9J9 A0A2J7PND9 A0A1Y1MD58 W5JB70 A0A1I8JTM0 A0A182L7Y4 A0A182ILT2 Q7Q110 A0A2M4BH96 A0A023EVD8 A0A2M3Z2M0 A0A2M4AA45 A0A2M4BHC9 A0A182UZX6 Q171C2 A0A182QJ54 B0WGC1 U5EWA6 A0A182MJF2 A0A1Q3FBL3 A0A0L0CIX9 A0A1B0GBD0 A0A1B0A641 A0A1A9WU99 A0A0K8VPA2 W8C540 A0A0K8V746 A0A182PEF7 A0A0A1XKK7 A0A1B0DH71 A0A1I8NQN6 A0A0M3QU86 A0A1B0CTY3 B4M8K2 Q9V3N6 B4KIW7 A0A1L8DUL4 A0A195BYT1 A0A158NMA9 A0A182F339 A0A1I8M8T6 A0A3B0JYF3 A0A0J9R241 A0A151JVR9 B4HX86 A0A1W4V7D4 A0A151IE66 B4P3P2 A0A0J7L1M0 B4MYQ8 F4WK19 A0A3L8DL30 Q29K63 B4GWY4 A0A026WFE1 B3NAE5 A0A1A9V1P4 A0A151X279 A0A154P6B9 A0A088A1M4 A0A2A3EJ76 A0A0C9Q6Y4 A0A0L7QVT0 A0A139WH92 E9J4J5 B4JCW2 B3MV54 A0A1W4WPF0 A0A182TEF5 A0A1B6K137 B4Q550 E2C2S3 A0A1B6ETI8 A0A1B6LF31 A0A0L7LKQ9 A0A1B6DT05 K7IQV5 A0A182NNY2 A0A310SIS0 A0A1W7R9E1 A0A2L2YDS0 A0A182R1U2 A0A182W8M6 A0A2L2YDT7 L7M2F2 A0A3B0JWJ9 A0A131YHE9 A0A224ZAY0 A0A1B0C3N8 A0A182GUY6 A0A0P5IPU6 A0A0P6BYV3 E0VU80 A0A3B0JTL8
A0A067R9J9 A0A2J7PND9 A0A1Y1MD58 W5JB70 A0A1I8JTM0 A0A182L7Y4 A0A182ILT2 Q7Q110 A0A2M4BH96 A0A023EVD8 A0A2M3Z2M0 A0A2M4AA45 A0A2M4BHC9 A0A182UZX6 Q171C2 A0A182QJ54 B0WGC1 U5EWA6 A0A182MJF2 A0A1Q3FBL3 A0A0L0CIX9 A0A1B0GBD0 A0A1B0A641 A0A1A9WU99 A0A0K8VPA2 W8C540 A0A0K8V746 A0A182PEF7 A0A0A1XKK7 A0A1B0DH71 A0A1I8NQN6 A0A0M3QU86 A0A1B0CTY3 B4M8K2 Q9V3N6 B4KIW7 A0A1L8DUL4 A0A195BYT1 A0A158NMA9 A0A182F339 A0A1I8M8T6 A0A3B0JYF3 A0A0J9R241 A0A151JVR9 B4HX86 A0A1W4V7D4 A0A151IE66 B4P3P2 A0A0J7L1M0 B4MYQ8 F4WK19 A0A3L8DL30 Q29K63 B4GWY4 A0A026WFE1 B3NAE5 A0A1A9V1P4 A0A151X279 A0A154P6B9 A0A088A1M4 A0A2A3EJ76 A0A0C9Q6Y4 A0A0L7QVT0 A0A139WH92 E9J4J5 B4JCW2 B3MV54 A0A1W4WPF0 A0A182TEF5 A0A1B6K137 B4Q550 E2C2S3 A0A1B6ETI8 A0A1B6LF31 A0A0L7LKQ9 A0A1B6DT05 K7IQV5 A0A182NNY2 A0A310SIS0 A0A1W7R9E1 A0A2L2YDS0 A0A182R1U2 A0A182W8M6 A0A2L2YDT7 L7M2F2 A0A3B0JWJ9 A0A131YHE9 A0A224ZAY0 A0A1B0C3N8 A0A182GUY6 A0A0P5IPU6 A0A0P6BYV3 E0VU80 A0A3B0JTL8
Pubmed
19121390
22118469
26354079
24845553
28004739
20920257
+ More
23761445 20966253 12364791 24945155 17510324 26108605 24495485 25830018 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 25315136 22936249 17550304 21719571 30249741 15632085 24508170 18362917 19820115 21282665 20798317 26227816 20075255 26561354 25576852 26830274 28797301 26483478 20566863
23761445 20966253 12364791 24945155 17510324 26108605 24495485 25830018 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 25315136 22936249 17550304 21719571 30249741 15632085 24508170 18362917 19820115 21282665 20798317 26227816 20075255 26561354 25576852 26830274 28797301 26483478 20566863
EMBL
BABH01010518
ODYU01011824
SOQ57786.1
AGBW02009105
OWR51676.1
KQ461196
+ More
KPJ06480.1 GDQN01006454 JAT84600.1 KQ459601 KPI93722.1 KK852840 KDR15208.1 NEVH01023954 PNF17845.1 GEZM01039708 JAV81277.1 ADMH02001979 ETN60095.1 APCN01001645 AAAB01008980 EAA13912.2 GGFJ01003180 MBW52321.1 GAPW01000500 JAC13098.1 GGFM01002013 MBW22764.1 GGFK01004353 MBW37674.1 GGFJ01003220 MBW52361.1 CH477459 EAT40604.1 AXCN02002097 DS231924 EDS26859.1 GANO01001505 JAB58366.1 AXCM01001780 GFDL01010157 JAV24888.1 JRES01000422 KNC31414.1 CCAG010022546 GDHF01011636 JAI40678.1 GAMC01001982 JAC04574.1 GDHF01017585 JAI34729.1 GBXI01002751 JAD11541.1 AJVK01006029 AJVK01006030 CP012523 ALC40199.1 AJWK01028170 AJWK01028171 CH940654 EDW57528.1 AE014134 AY058275 AAF53324.1 AAL13504.1 AHN54421.1 CH933807 EDW13480.1 GFDF01003952 JAV10132.1 KQ976395 KYM93086.1 ADTU01020321 OUUW01000010 SPP85462.1 CM002910 KMY90148.1 KQ981697 KYN37537.1 CH480818 EDW51666.1 KQ977890 KYM98989.1 CM000157 EDW88343.1 LBMM01001235 KMQ96672.1 CH963913 EDW77247.1 GL888190 EGI65469.1 QOIP01000007 RLU21081.1 CH379061 EAL33315.2 CH479195 EDW27311.1 KK107238 EZA54777.1 CH954177 EDV58647.1 KQ982584 KYQ54398.1 KQ434819 KZC06894.1 KZ288250 PBC31081.1 GBYB01009878 JAG79645.1 KQ414722 KOC62710.1 KQ971343 KYB27333.1 GL768105 EFZ12247.1 CH916368 EDW03201.1 CH902624 EDV33119.1 GECU01002539 JAT05168.1 CM000361 EDX04956.1 GL452203 EFN77758.1 GECZ01028569 JAS41200.1 GEBQ01017672 JAT22305.1 JTDY01000815 KOB75776.1 GEDC01028677 GEDC01008498 GEDC01000983 JAS08621.1 JAS28800.1 JAS36315.1 KQ765234 OAD54014.1 GFAH01000628 JAV47761.1 IAAA01010797 LAA06278.1 IAAA01010798 LAA06279.1 GACK01006819 JAA58215.1 SPP85463.1 GEDV01010200 JAP78357.1 GFPF01012766 MAA23912.1 JXJN01025052 JXUM01089932 JXUM01089933 JXUM01089934 KQ563805 KXJ73252.1 GDIQ01210399 JAK41326.1 GDIP01007342 LRGB01000024 JAM96373.1 KZS21374.1 DS235781 EEB16936.1 SPP85464.1
KPJ06480.1 GDQN01006454 JAT84600.1 KQ459601 KPI93722.1 KK852840 KDR15208.1 NEVH01023954 PNF17845.1 GEZM01039708 JAV81277.1 ADMH02001979 ETN60095.1 APCN01001645 AAAB01008980 EAA13912.2 GGFJ01003180 MBW52321.1 GAPW01000500 JAC13098.1 GGFM01002013 MBW22764.1 GGFK01004353 MBW37674.1 GGFJ01003220 MBW52361.1 CH477459 EAT40604.1 AXCN02002097 DS231924 EDS26859.1 GANO01001505 JAB58366.1 AXCM01001780 GFDL01010157 JAV24888.1 JRES01000422 KNC31414.1 CCAG010022546 GDHF01011636 JAI40678.1 GAMC01001982 JAC04574.1 GDHF01017585 JAI34729.1 GBXI01002751 JAD11541.1 AJVK01006029 AJVK01006030 CP012523 ALC40199.1 AJWK01028170 AJWK01028171 CH940654 EDW57528.1 AE014134 AY058275 AAF53324.1 AAL13504.1 AHN54421.1 CH933807 EDW13480.1 GFDF01003952 JAV10132.1 KQ976395 KYM93086.1 ADTU01020321 OUUW01000010 SPP85462.1 CM002910 KMY90148.1 KQ981697 KYN37537.1 CH480818 EDW51666.1 KQ977890 KYM98989.1 CM000157 EDW88343.1 LBMM01001235 KMQ96672.1 CH963913 EDW77247.1 GL888190 EGI65469.1 QOIP01000007 RLU21081.1 CH379061 EAL33315.2 CH479195 EDW27311.1 KK107238 EZA54777.1 CH954177 EDV58647.1 KQ982584 KYQ54398.1 KQ434819 KZC06894.1 KZ288250 PBC31081.1 GBYB01009878 JAG79645.1 KQ414722 KOC62710.1 KQ971343 KYB27333.1 GL768105 EFZ12247.1 CH916368 EDW03201.1 CH902624 EDV33119.1 GECU01002539 JAT05168.1 CM000361 EDX04956.1 GL452203 EFN77758.1 GECZ01028569 JAS41200.1 GEBQ01017672 JAT22305.1 JTDY01000815 KOB75776.1 GEDC01028677 GEDC01008498 GEDC01000983 JAS08621.1 JAS28800.1 JAS36315.1 KQ765234 OAD54014.1 GFAH01000628 JAV47761.1 IAAA01010797 LAA06278.1 IAAA01010798 LAA06279.1 GACK01006819 JAA58215.1 SPP85463.1 GEDV01010200 JAP78357.1 GFPF01012766 MAA23912.1 JXJN01025052 JXUM01089932 JXUM01089933 JXUM01089934 KQ563805 KXJ73252.1 GDIQ01210399 JAK41326.1 GDIP01007342 LRGB01000024 JAM96373.1 KZS21374.1 DS235781 EEB16936.1 SPP85464.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000027135
UP000235965
+ More
UP000000673 UP000075840 UP000075882 UP000075880 UP000007062 UP000075903 UP000008820 UP000075886 UP000002320 UP000075883 UP000037069 UP000092444 UP000092445 UP000091820 UP000075885 UP000092462 UP000095300 UP000092553 UP000092461 UP000008792 UP000000803 UP000009192 UP000078540 UP000005205 UP000069272 UP000095301 UP000268350 UP000078541 UP000001292 UP000192221 UP000078542 UP000002282 UP000036403 UP000007798 UP000007755 UP000279307 UP000001819 UP000008744 UP000053097 UP000008711 UP000078200 UP000075809 UP000076502 UP000005203 UP000242457 UP000053825 UP000007266 UP000001070 UP000007801 UP000192223 UP000075902 UP000000304 UP000008237 UP000037510 UP000002358 UP000075884 UP000075900 UP000075920 UP000092460 UP000069940 UP000249989 UP000076858 UP000009046
UP000000673 UP000075840 UP000075882 UP000075880 UP000007062 UP000075903 UP000008820 UP000075886 UP000002320 UP000075883 UP000037069 UP000092444 UP000092445 UP000091820 UP000075885 UP000092462 UP000095300 UP000092553 UP000092461 UP000008792 UP000000803 UP000009192 UP000078540 UP000005205 UP000069272 UP000095301 UP000268350 UP000078541 UP000001292 UP000192221 UP000078542 UP000002282 UP000036403 UP000007798 UP000007755 UP000279307 UP000001819 UP000008744 UP000053097 UP000008711 UP000078200 UP000075809 UP000076502 UP000005203 UP000242457 UP000053825 UP000007266 UP000001070 UP000007801 UP000192223 UP000075902 UP000000304 UP000008237 UP000037510 UP000002358 UP000075884 UP000075900 UP000075920 UP000092460 UP000069940 UP000249989 UP000076858 UP000009046
Pfam
Interpro
IPR004240
EMP70
+ More
IPR010666 Znf_GRF
IPR013825 Topo_IA_cen_sub2
IPR003602 Topo_IA_DNA-bd_dom
IPR020846 MFS_dom
IPR023405 Topo_IA_core_domain
IPR013498 Topo_IA_Znf
IPR023406 Topo_IA_AS
IPR034144 TOPRIM_TopoIII
IPR013824 Topo_IA_cen_sub1
IPR003601 Topo_IA_2
IPR013826 Topo_IA_cen_sub3
IPR013497 Topo_IA_cen
IPR006171 TOPRIM_domain
IPR023298 ATPase_P-typ_TM_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR019775 WD40_repeat_CS
IPR038184 CSTF1_dimer_sf
IPR036322 WD40_repeat_dom_sf
IPR032028 CSTF1_dimer
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
IPR010666 Znf_GRF
IPR013825 Topo_IA_cen_sub2
IPR003602 Topo_IA_DNA-bd_dom
IPR020846 MFS_dom
IPR023405 Topo_IA_core_domain
IPR013498 Topo_IA_Znf
IPR023406 Topo_IA_AS
IPR034144 TOPRIM_TopoIII
IPR013824 Topo_IA_cen_sub1
IPR003601 Topo_IA_2
IPR013826 Topo_IA_cen_sub3
IPR013497 Topo_IA_cen
IPR006171 TOPRIM_domain
IPR023298 ATPase_P-typ_TM_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR017986 WD40_repeat_dom
IPR001680 WD40_repeat
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR019775 WD40_repeat_CS
IPR038184 CSTF1_dimer_sf
IPR036322 WD40_repeat_dom_sf
IPR032028 CSTF1_dimer
IPR036291 NAD(P)-bd_dom_sf
IPR002347 SDR_fam
SUPFAM
ProteinModelPortal
H9J0E2
A0A2H1WXU4
A0A212FD46
A0A194QND0
A0A1E1WC95
A0A194PK74
+ More
A0A067R9J9 A0A2J7PND9 A0A1Y1MD58 W5JB70 A0A1I8JTM0 A0A182L7Y4 A0A182ILT2 Q7Q110 A0A2M4BH96 A0A023EVD8 A0A2M3Z2M0 A0A2M4AA45 A0A2M4BHC9 A0A182UZX6 Q171C2 A0A182QJ54 B0WGC1 U5EWA6 A0A182MJF2 A0A1Q3FBL3 A0A0L0CIX9 A0A1B0GBD0 A0A1B0A641 A0A1A9WU99 A0A0K8VPA2 W8C540 A0A0K8V746 A0A182PEF7 A0A0A1XKK7 A0A1B0DH71 A0A1I8NQN6 A0A0M3QU86 A0A1B0CTY3 B4M8K2 Q9V3N6 B4KIW7 A0A1L8DUL4 A0A195BYT1 A0A158NMA9 A0A182F339 A0A1I8M8T6 A0A3B0JYF3 A0A0J9R241 A0A151JVR9 B4HX86 A0A1W4V7D4 A0A151IE66 B4P3P2 A0A0J7L1M0 B4MYQ8 F4WK19 A0A3L8DL30 Q29K63 B4GWY4 A0A026WFE1 B3NAE5 A0A1A9V1P4 A0A151X279 A0A154P6B9 A0A088A1M4 A0A2A3EJ76 A0A0C9Q6Y4 A0A0L7QVT0 A0A139WH92 E9J4J5 B4JCW2 B3MV54 A0A1W4WPF0 A0A182TEF5 A0A1B6K137 B4Q550 E2C2S3 A0A1B6ETI8 A0A1B6LF31 A0A0L7LKQ9 A0A1B6DT05 K7IQV5 A0A182NNY2 A0A310SIS0 A0A1W7R9E1 A0A2L2YDS0 A0A182R1U2 A0A182W8M6 A0A2L2YDT7 L7M2F2 A0A3B0JWJ9 A0A131YHE9 A0A224ZAY0 A0A1B0C3N8 A0A182GUY6 A0A0P5IPU6 A0A0P6BYV3 E0VU80 A0A3B0JTL8
A0A067R9J9 A0A2J7PND9 A0A1Y1MD58 W5JB70 A0A1I8JTM0 A0A182L7Y4 A0A182ILT2 Q7Q110 A0A2M4BH96 A0A023EVD8 A0A2M3Z2M0 A0A2M4AA45 A0A2M4BHC9 A0A182UZX6 Q171C2 A0A182QJ54 B0WGC1 U5EWA6 A0A182MJF2 A0A1Q3FBL3 A0A0L0CIX9 A0A1B0GBD0 A0A1B0A641 A0A1A9WU99 A0A0K8VPA2 W8C540 A0A0K8V746 A0A182PEF7 A0A0A1XKK7 A0A1B0DH71 A0A1I8NQN6 A0A0M3QU86 A0A1B0CTY3 B4M8K2 Q9V3N6 B4KIW7 A0A1L8DUL4 A0A195BYT1 A0A158NMA9 A0A182F339 A0A1I8M8T6 A0A3B0JYF3 A0A0J9R241 A0A151JVR9 B4HX86 A0A1W4V7D4 A0A151IE66 B4P3P2 A0A0J7L1M0 B4MYQ8 F4WK19 A0A3L8DL30 Q29K63 B4GWY4 A0A026WFE1 B3NAE5 A0A1A9V1P4 A0A151X279 A0A154P6B9 A0A088A1M4 A0A2A3EJ76 A0A0C9Q6Y4 A0A0L7QVT0 A0A139WH92 E9J4J5 B4JCW2 B3MV54 A0A1W4WPF0 A0A182TEF5 A0A1B6K137 B4Q550 E2C2S3 A0A1B6ETI8 A0A1B6LF31 A0A0L7LKQ9 A0A1B6DT05 K7IQV5 A0A182NNY2 A0A310SIS0 A0A1W7R9E1 A0A2L2YDS0 A0A182R1U2 A0A182W8M6 A0A2L2YDT7 L7M2F2 A0A3B0JWJ9 A0A131YHE9 A0A224ZAY0 A0A1B0C3N8 A0A182GUY6 A0A0P5IPU6 A0A0P6BYV3 E0VU80 A0A3B0JTL8
Ontologies
GO
PANTHER
Topology
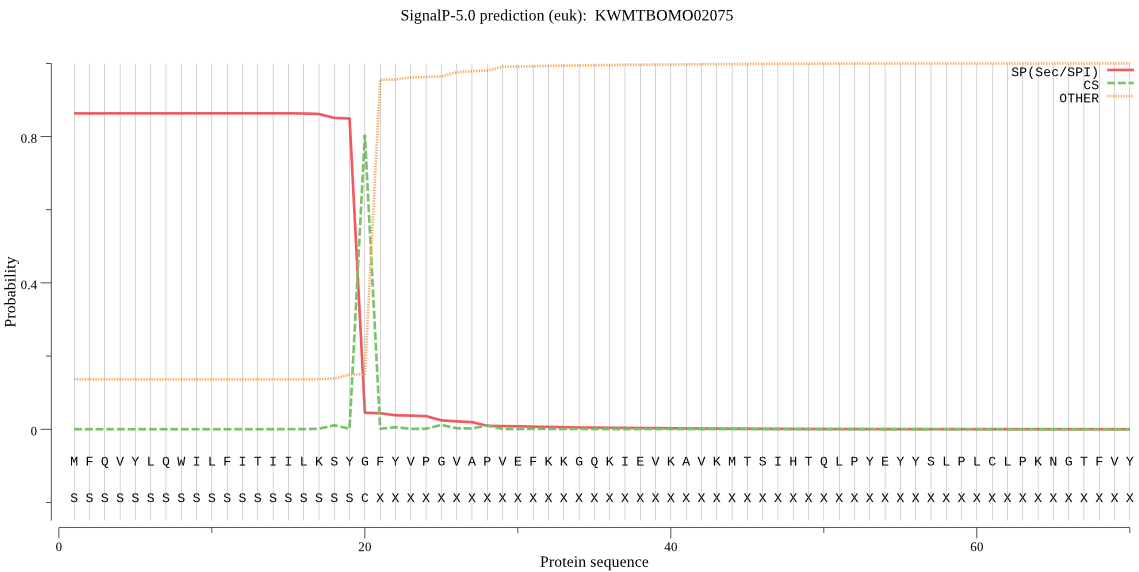
SignalP
Position: 1 - 20,
Likelihood: 0.862654
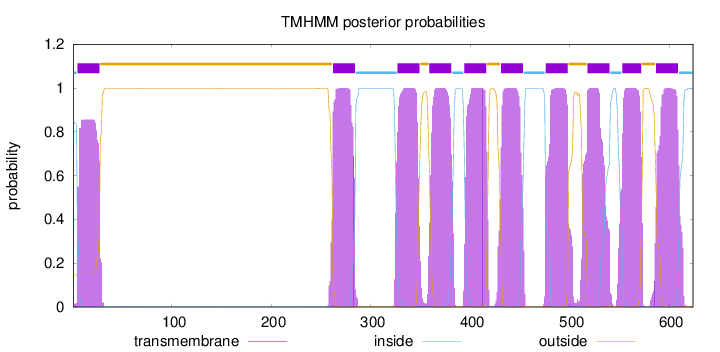
Length:
624
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
223.50593
Exp number, first 60 AAs:
19.06029
Total prob of N-in:
0.85256
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 261
TMhelix
262 - 284
inside
285 - 326
TMhelix
327 - 349
outside
350 - 358
TMhelix
359 - 381
inside
382 - 393
TMhelix
394 - 416
outside
417 - 430
TMhelix
431 - 453
inside
454 - 475
TMhelix
476 - 498
outside
499 - 517
TMhelix
518 - 540
inside
541 - 552
TMhelix
553 - 572
outside
573 - 586
TMhelix
587 - 609
inside
610 - 624
Population Genetic Test Statistics
Pi
292.497831
Theta
207.505331
Tajima's D
1.156993
CLR
0.046175
CSRT
0.697315134243288
Interpretation
Uncertain
