Pre Gene Modal
BGIBMGA003191
Annotation
PREDICTED:_neuroguidin_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 4.423
Sequence
CDS
ATGGTCCAGGCCTGCGAAGAAACAGAACAACCAAACATCCCTCAAGCAATGAAATTATTAAAAGAAATGAATGTAAATGTACAACAAGTATCTCAGTTAGTCGATAACATGCTTGTCCGTGTGAAGAATGGAGAACTTTCAACAGACAAAGGCCTTAGCTTTTTGGAAATGAAATACCAAATGCTTCTTAGTTACTTGATCAATTTGACCTATATAGTATTAAGGAAGTGCTCTGGTGAAAGAATTGAATCTGATCCTTCCATTGATAGGCTCATTGAAATAAGAACAGTACTTGAAAAGATTCGTCCAATTGATTCTAAATTAAAGTACCAAATTGATAAGCTTGTCAAAAGTGCAGTTGTTGGAGCTGTATCAGAAGATGACCCACAAACCTACCGTGCTAATCCTAATAACTTAGTAAGTAAGATTGATAATTCAGAAGAAGAGTCAAGTGAAACCTCTGATGTAGAAGGCAAAAAAGAAAAAGTAAAATCTAATATTTATGTGCCTCCAAAATTAGCTGCAGTACATTTTGATGAAGTCTCAAGAACTGAAGCAGATAAAAAAATTAAAGAGAAATCGAAGAAACAATTTCTCAATTCCAGTGTTATGAGGGAACTTCGAGAAGAATATTCAGAAGCACCATTAGAAGTCAGCTCCGGGAATCATATTAAACAATCCATATCCAAATATGAACAGGAAAAAACTGAATATGAAGAAAATTATCTCACAAGATTACCTGTAACAAAAGCTGAAAAAAATAGGAGAAAAAAAGTAACCACAGTAGGGATGTTGGCTGATGAGATAACAGGCACAAGTAGTCAAAGAGTAGCTCGAAAACACAAAATGAAATCTAAAAAAGGAAAAGGATTCAAGAGAAGGCGTATACATTGA
Protein
MVQACEETEQPNIPQAMKLLKEMNVNVQQVSQLVDNMLVRVKNGELSTDKGLSFLEMKYQMLLSYLINLTYIVLRKCSGERIESDPSIDRLIEIRTVLEKIRPIDSKLKYQIDKLVKSAVVGAVSEDDPQTYRANPNNLVSKIDNSEEESSETSDVEGKKEKVKSNIYVPPKLAAVHFDEVSRTEADKKIKEKSKKQFLNSSVMRELREEYSEAPLEVSSGNHIKQSISKYEQEKTEYEENYLTRLPVTKAEKNRRKKVTTVGMLADEITGTSSQRVARKHKMKSKKGKGFKRRRIH
Summary
Uniprot
H9J104
A0A2H1WXP5
A0A194PL47
A0A194QSG2
A0A0L7LK97
A0A1E1VYR9
+ More
A0A212FD85 A0A0J7L248 F4WK20 A0A158NM69 E2C2S2 E9J4J6 V9IHE6 A0A088A1E4 A0A232FCJ8 A0A195BXR5 A0A0N1ITT9 A0A0C9PXX2 K7IQU7 A0A2A3EH88 A0A151ISI7 A0A151X2H7 A0A151JVM9 E2ABV8 A0A026WGF4 A0A154P4W1 A0A0L7QVY8 A0A310SAK2 A0A151IE57 A0A2J7PNE9 A0A1Y1M963 A0A067QXN8 A0A0T6B085 A0A1W4X3F6 A0A3L8DLF0 T1PLE8 A0A1I8N0Y0 D6WLZ7 A0A0K8UTW4 A0A034WKR8 B4KGZ6 A0A1I8P7U2 A0A1A9X060 A0A1I8P815 A0A0J9QXB2 A0A0A1XFD7 B3MMM0 Q9VMM7 W8C8C1 Q8IGY8 Q29PJ7 B3N571 A0A1W4V500 B4LRJ6 A0A1Q3EWC6 A0A1Q3EW28 B4P030 A0A1Q3EW31 A0A3B0JVB5 B4JC68 A0A0M3QTF6 A0A336LWY1 A0A0L0BX31 B4MV28 A0A3B0KCT8 A0A3B0KQ93 Q16YR9 A0A182QZ04 A0A182G3M5 B0W0H6 A0A182NAK2 A0A1I8P7Z0 B4Q423 A0A1W4V3U8 A0A182YR68 A0A1J1I1X7 N6TBY0 U4U5G2 N6T4Z6 B4G6W1 A0A182UG58 A0A0K8TRQ1 A0A1L8DGZ7 A0A182SA71 A0A1B6CDM5 T1GCF7 A0A1B0A3U3 A0A1B0ANU5 A0A1B0D5Z0 T1JJK7 A0A1B6H1S0 A0A069DR66 A0A224XU94 A0A0P4VIA1 A0A1B6IE43 A0A023F0U8 A0A1A9YNR7
A0A212FD85 A0A0J7L248 F4WK20 A0A158NM69 E2C2S2 E9J4J6 V9IHE6 A0A088A1E4 A0A232FCJ8 A0A195BXR5 A0A0N1ITT9 A0A0C9PXX2 K7IQU7 A0A2A3EH88 A0A151ISI7 A0A151X2H7 A0A151JVM9 E2ABV8 A0A026WGF4 A0A154P4W1 A0A0L7QVY8 A0A310SAK2 A0A151IE57 A0A2J7PNE9 A0A1Y1M963 A0A067QXN8 A0A0T6B085 A0A1W4X3F6 A0A3L8DLF0 T1PLE8 A0A1I8N0Y0 D6WLZ7 A0A0K8UTW4 A0A034WKR8 B4KGZ6 A0A1I8P7U2 A0A1A9X060 A0A1I8P815 A0A0J9QXB2 A0A0A1XFD7 B3MMM0 Q9VMM7 W8C8C1 Q8IGY8 Q29PJ7 B3N571 A0A1W4V500 B4LRJ6 A0A1Q3EWC6 A0A1Q3EW28 B4P030 A0A1Q3EW31 A0A3B0JVB5 B4JC68 A0A0M3QTF6 A0A336LWY1 A0A0L0BX31 B4MV28 A0A3B0KCT8 A0A3B0KQ93 Q16YR9 A0A182QZ04 A0A182G3M5 B0W0H6 A0A182NAK2 A0A1I8P7Z0 B4Q423 A0A1W4V3U8 A0A182YR68 A0A1J1I1X7 N6TBY0 U4U5G2 N6T4Z6 B4G6W1 A0A182UG58 A0A0K8TRQ1 A0A1L8DGZ7 A0A182SA71 A0A1B6CDM5 T1GCF7 A0A1B0A3U3 A0A1B0ANU5 A0A1B0D5Z0 T1JJK7 A0A1B6H1S0 A0A069DR66 A0A224XU94 A0A0P4VIA1 A0A1B6IE43 A0A023F0U8 A0A1A9YNR7
Pubmed
19121390
26354079
26227816
22118469
21719571
21347285
+ More
20798317 21282665 28648823 20075255 24508170 28004739 24845553 30249741 25315136 18362917 19820115 25348373 17994087 22936249 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 17550304 26108605 17510324 26483478 25244985 23537049 26369729 26334808 27129103 25474469
20798317 21282665 28648823 20075255 24508170 28004739 24845553 30249741 25315136 18362917 19820115 25348373 17994087 22936249 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 15632085 17550304 26108605 17510324 26483478 25244985 23537049 26369729 26334808 27129103 25474469
EMBL
BABH01010518
ODYU01011824
SOQ57787.1
KQ459601
KPI93723.1
KQ461196
+ More
KPJ06481.1 JTDY01000815 KOB75779.1 GDQN01011225 JAT79829.1 AGBW02009105 OWR51677.1 LBMM01001235 KMQ96673.1 GL888190 EGI65470.1 ADTU01020319 GL452203 EFN77757.1 GL768105 EFZ12244.1 JR047677 AEY60480.1 NNAY01000461 OXU28210.1 KQ976395 KYM93085.1 KQ435748 KOX76408.1 GBYB01006353 JAG76120.1 KZ288250 PBC31080.1 KQ981090 KYN09574.1 KQ982584 KYQ54397.1 KQ981697 KYN37536.1 GL438386 EFN69053.1 KK107238 EZA54776.1 KQ434819 KZC06893.1 KQ414722 KOC62709.1 KQ765234 OAD54013.1 KQ977890 KYM98988.1 NEVH01023954 PNF17839.1 GEZM01036916 JAV82342.1 KK852840 KDR15210.1 LJIG01016375 KRT80790.1 QOIP01000007 RLU21033.1 KA648920 AFP63549.1 KQ971343 EFA04194.1 GDHF01022278 JAI30036.1 GAKP01004242 JAC54710.1 CH933807 EDW12207.2 CM002910 KMY88359.1 GBXI01006718 GBXI01004178 JAD07574.1 JAD10114.1 CH902620 EDV31911.2 AE014134 BT046133 AAF52286.2 ACI46521.2 GAMC01007561 JAB98994.1 BT001525 AAN71280.1 CH379058 EAL34296.3 CH954177 EDV57901.2 CH940649 EDW63591.2 GFDL01015537 JAV19508.1 GFDL01015533 JAV19512.1 CM000157 EDW88895.2 GFDL01015530 JAV19515.1 OUUW01000010 SPP86035.1 CH916368 EDW03081.1 CP012523 ALC38799.1 UFQT01000144 SSX20817.1 JRES01001211 KNC24556.1 CH963857 EDW76373.2 SPP86040.1 SPP86038.1 CH477510 EAT39780.1 AXCN02002191 JXUM01041044 KQ561268 KXJ79149.1 DS231817 EDS39715.1 CM000361 EDX03856.1 CVRI01000038 CRK93762.1 APGK01043875 KB741018 ENN75238.1 KB632152 ERL89134.1 ENN75239.1 CH479180 EDW28280.1 GDAI01000785 JAI16818.1 GFDF01008368 JAV05716.1 GEDC01025767 JAS11531.1 CAQQ02176272 CAQQ02176273 JXJN01001022 AJVK01003472 JH431850 GECZ01001137 JAS68632.1 GBGD01002321 JAC86568.1 GFTR01004753 JAW11673.1 GDKW01002129 JAI54466.1 GECU01022534 JAS85172.1 GBBI01003607 JAC15105.1
KPJ06481.1 JTDY01000815 KOB75779.1 GDQN01011225 JAT79829.1 AGBW02009105 OWR51677.1 LBMM01001235 KMQ96673.1 GL888190 EGI65470.1 ADTU01020319 GL452203 EFN77757.1 GL768105 EFZ12244.1 JR047677 AEY60480.1 NNAY01000461 OXU28210.1 KQ976395 KYM93085.1 KQ435748 KOX76408.1 GBYB01006353 JAG76120.1 KZ288250 PBC31080.1 KQ981090 KYN09574.1 KQ982584 KYQ54397.1 KQ981697 KYN37536.1 GL438386 EFN69053.1 KK107238 EZA54776.1 KQ434819 KZC06893.1 KQ414722 KOC62709.1 KQ765234 OAD54013.1 KQ977890 KYM98988.1 NEVH01023954 PNF17839.1 GEZM01036916 JAV82342.1 KK852840 KDR15210.1 LJIG01016375 KRT80790.1 QOIP01000007 RLU21033.1 KA648920 AFP63549.1 KQ971343 EFA04194.1 GDHF01022278 JAI30036.1 GAKP01004242 JAC54710.1 CH933807 EDW12207.2 CM002910 KMY88359.1 GBXI01006718 GBXI01004178 JAD07574.1 JAD10114.1 CH902620 EDV31911.2 AE014134 BT046133 AAF52286.2 ACI46521.2 GAMC01007561 JAB98994.1 BT001525 AAN71280.1 CH379058 EAL34296.3 CH954177 EDV57901.2 CH940649 EDW63591.2 GFDL01015537 JAV19508.1 GFDL01015533 JAV19512.1 CM000157 EDW88895.2 GFDL01015530 JAV19515.1 OUUW01000010 SPP86035.1 CH916368 EDW03081.1 CP012523 ALC38799.1 UFQT01000144 SSX20817.1 JRES01001211 KNC24556.1 CH963857 EDW76373.2 SPP86040.1 SPP86038.1 CH477510 EAT39780.1 AXCN02002191 JXUM01041044 KQ561268 KXJ79149.1 DS231817 EDS39715.1 CM000361 EDX03856.1 CVRI01000038 CRK93762.1 APGK01043875 KB741018 ENN75238.1 KB632152 ERL89134.1 ENN75239.1 CH479180 EDW28280.1 GDAI01000785 JAI16818.1 GFDF01008368 JAV05716.1 GEDC01025767 JAS11531.1 CAQQ02176272 CAQQ02176273 JXJN01001022 AJVK01003472 JH431850 GECZ01001137 JAS68632.1 GBGD01002321 JAC86568.1 GFTR01004753 JAW11673.1 GDKW01002129 JAI54466.1 GECU01022534 JAS85172.1 GBBI01003607 JAC15105.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000036403
+ More
UP000007755 UP000005205 UP000008237 UP000005203 UP000215335 UP000078540 UP000053105 UP000002358 UP000242457 UP000078492 UP000075809 UP000078541 UP000000311 UP000053097 UP000076502 UP000053825 UP000078542 UP000235965 UP000027135 UP000192223 UP000279307 UP000095301 UP000007266 UP000009192 UP000095300 UP000091820 UP000007801 UP000000803 UP000001819 UP000008711 UP000192221 UP000008792 UP000002282 UP000268350 UP000001070 UP000092553 UP000037069 UP000007798 UP000008820 UP000075886 UP000069940 UP000249989 UP000002320 UP000075884 UP000000304 UP000076408 UP000183832 UP000019118 UP000030742 UP000008744 UP000075902 UP000075901 UP000015102 UP000092445 UP000092460 UP000092462 UP000092443
UP000007755 UP000005205 UP000008237 UP000005203 UP000215335 UP000078540 UP000053105 UP000002358 UP000242457 UP000078492 UP000075809 UP000078541 UP000000311 UP000053097 UP000076502 UP000053825 UP000078542 UP000235965 UP000027135 UP000192223 UP000279307 UP000095301 UP000007266 UP000009192 UP000095300 UP000091820 UP000007801 UP000000803 UP000001819 UP000008711 UP000192221 UP000008792 UP000002282 UP000268350 UP000001070 UP000092553 UP000037069 UP000007798 UP000008820 UP000075886 UP000069940 UP000249989 UP000002320 UP000075884 UP000000304 UP000076408 UP000183832 UP000019118 UP000030742 UP000008744 UP000075902 UP000075901 UP000015102 UP000092445 UP000092460 UP000092462 UP000092443
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9J104
A0A2H1WXP5
A0A194PL47
A0A194QSG2
A0A0L7LK97
A0A1E1VYR9
+ More
A0A212FD85 A0A0J7L248 F4WK20 A0A158NM69 E2C2S2 E9J4J6 V9IHE6 A0A088A1E4 A0A232FCJ8 A0A195BXR5 A0A0N1ITT9 A0A0C9PXX2 K7IQU7 A0A2A3EH88 A0A151ISI7 A0A151X2H7 A0A151JVM9 E2ABV8 A0A026WGF4 A0A154P4W1 A0A0L7QVY8 A0A310SAK2 A0A151IE57 A0A2J7PNE9 A0A1Y1M963 A0A067QXN8 A0A0T6B085 A0A1W4X3F6 A0A3L8DLF0 T1PLE8 A0A1I8N0Y0 D6WLZ7 A0A0K8UTW4 A0A034WKR8 B4KGZ6 A0A1I8P7U2 A0A1A9X060 A0A1I8P815 A0A0J9QXB2 A0A0A1XFD7 B3MMM0 Q9VMM7 W8C8C1 Q8IGY8 Q29PJ7 B3N571 A0A1W4V500 B4LRJ6 A0A1Q3EWC6 A0A1Q3EW28 B4P030 A0A1Q3EW31 A0A3B0JVB5 B4JC68 A0A0M3QTF6 A0A336LWY1 A0A0L0BX31 B4MV28 A0A3B0KCT8 A0A3B0KQ93 Q16YR9 A0A182QZ04 A0A182G3M5 B0W0H6 A0A182NAK2 A0A1I8P7Z0 B4Q423 A0A1W4V3U8 A0A182YR68 A0A1J1I1X7 N6TBY0 U4U5G2 N6T4Z6 B4G6W1 A0A182UG58 A0A0K8TRQ1 A0A1L8DGZ7 A0A182SA71 A0A1B6CDM5 T1GCF7 A0A1B0A3U3 A0A1B0ANU5 A0A1B0D5Z0 T1JJK7 A0A1B6H1S0 A0A069DR66 A0A224XU94 A0A0P4VIA1 A0A1B6IE43 A0A023F0U8 A0A1A9YNR7
A0A212FD85 A0A0J7L248 F4WK20 A0A158NM69 E2C2S2 E9J4J6 V9IHE6 A0A088A1E4 A0A232FCJ8 A0A195BXR5 A0A0N1ITT9 A0A0C9PXX2 K7IQU7 A0A2A3EH88 A0A151ISI7 A0A151X2H7 A0A151JVM9 E2ABV8 A0A026WGF4 A0A154P4W1 A0A0L7QVY8 A0A310SAK2 A0A151IE57 A0A2J7PNE9 A0A1Y1M963 A0A067QXN8 A0A0T6B085 A0A1W4X3F6 A0A3L8DLF0 T1PLE8 A0A1I8N0Y0 D6WLZ7 A0A0K8UTW4 A0A034WKR8 B4KGZ6 A0A1I8P7U2 A0A1A9X060 A0A1I8P815 A0A0J9QXB2 A0A0A1XFD7 B3MMM0 Q9VMM7 W8C8C1 Q8IGY8 Q29PJ7 B3N571 A0A1W4V500 B4LRJ6 A0A1Q3EWC6 A0A1Q3EW28 B4P030 A0A1Q3EW31 A0A3B0JVB5 B4JC68 A0A0M3QTF6 A0A336LWY1 A0A0L0BX31 B4MV28 A0A3B0KCT8 A0A3B0KQ93 Q16YR9 A0A182QZ04 A0A182G3M5 B0W0H6 A0A182NAK2 A0A1I8P7Z0 B4Q423 A0A1W4V3U8 A0A182YR68 A0A1J1I1X7 N6TBY0 U4U5G2 N6T4Z6 B4G6W1 A0A182UG58 A0A0K8TRQ1 A0A1L8DGZ7 A0A182SA71 A0A1B6CDM5 T1GCF7 A0A1B0A3U3 A0A1B0ANU5 A0A1B0D5Z0 T1JJK7 A0A1B6H1S0 A0A069DR66 A0A224XU94 A0A0P4VIA1 A0A1B6IE43 A0A023F0U8 A0A1A9YNR7
PDB
5WLC
E-value=0.0181744,
Score=88
Ontologies
GO
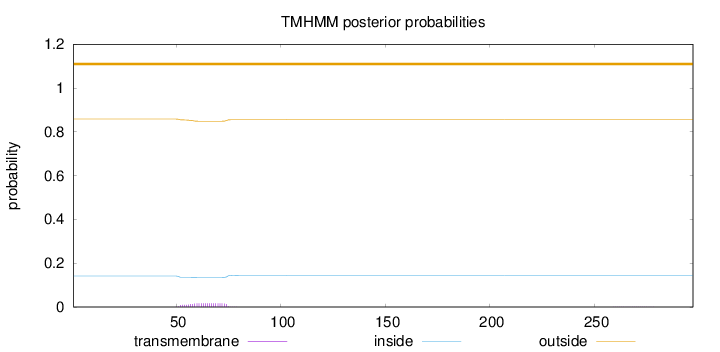
Topology
Length:
297
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.34974
Exp number, first 60 AAs:
0.11047
Total prob of N-in:
0.14164
outside
1 - 297
Population Genetic Test Statistics
Pi
274.152782
Theta
197.764622
Tajima's D
1.215757
CLR
0
CSRT
0.712914354282286
Interpretation
Uncertain
