Gene
KWMTBOMO02073
Annotation
PREDICTED:_radial_spoke_head_10_homolog_B-like_[Amyelois_transitella]
Full name
Radial spoke head 10 homolog B2
+ More
Radial spoke head 10 homolog B
Radial spoke head 10 homolog B
Location in the cell
Cytoplasmic Reliability : 2.189
Sequence
CDS
ATGTCTGGCTATGGTATTTACATTTGGGATGCCTATAATAATAATTCTTTATCTTTACCATCCATAAATATATACCGAGGCTTCTGGGTTAAGGGGCAGCGTAATGGCTACGGTATTTTGAATTTGGGGCTTGGCGCAAGTTCACACTACAGAGGAGAATTTCAAAATAACAAGAAGAACGGAACAGGAAAGTTTGTGACCAACAATGGCCTTGTTCTGCAGCATAAAACACTTTTTCTTAACGATAATTTAAGTGATATGGCGCCCGAGGAAGATTGTTGCGAATTGTTGCAACAAGAAAAACATTGTCAGGAAGTCGAACCTTTTTTATTCGATATATGTGCTGATCACACTATCGGACTTGCATACCATGTAGAAGAAGCTATAAAAAATATTGATAAAGGAATGGAAGACAGGAGTAAAATAATTAATGATTACTTGGAGAGTAACATGTCACAATACCGTAATGAGCTAAGGAGAAATACAATGCCTTACGACAATACTTACCCGTTCAGTATAGAAGACTTCATAGATTTTGAAGTAGATTCTCTACAAAAGTCACTTCGGTGTTATGAAACTGAGTTAAAAGTGATTTATTACAAGTATGCTACAATTTTCAATAAAGAGGAGATTAATTTCACACCGGTTTTAATACGATTTGGCTTGTGGCAATTATTTTTCGATTGCAATGTACACGAGAAGGGCCTAACACTTGTCGAAATCGATAAAATTTTCCACAAAAATCCTCAATGGTTAGCGAGAACACCACATAATCCTTTTGAAGAAATATATTTTTGGCAATTTCTACACAGTATATTATCTGTGGCCACTAAATTGTATGCATTGAAAGAGCTTCCTAAAACAAAACCAGACACGCTTCTGTCGACTGCGTTAAGACAGTTTATAGAAAAGGATGTTTGGCCAAATGTTGGTAGATGCAAAGGTCTGTCAGAGTAA
Protein
MSGYGIYIWDAYNNNSLSLPSINIYRGFWVKGQRNGYGILNLGLGASSHYRGEFQNNKKNGTGKFVTNNGLVLQHKTLFLNDNLSDMAPEEDCCELLQQEKHCQEVEPFLFDICADHTIGLAYHVEEAIKNIDKGMEDRSKIINDYLESNMSQYRNELRRNTMPYDNTYPFSIEDFIDFEVDSLQKSLRCYETELKVIYYKYATIFNKEEINFTPVLIRFGLWQLFFDCNVHEKGLTLVEIDKIFHKNPQWLARTPHNPFEEIYFWQFLHSILSVATKLYALKELPKTKPDTLLSTALRQFIEKDVWPNVGRCKGLSE
Summary
Keywords
Alternative splicing
Coiled coil
Complete proteome
Polymorphism
Reference proteome
Repeat
Feature
chain Radial spoke head 10 homolog B2
splice variant In isoform 2.
sequence variant In dbSNP:rs17855578.
splice variant In isoform 2.
sequence variant In dbSNP:rs17855578.
Uniprot
A0A212FD60
A0A0L7LK03
A0A194QLQ1
A0A194PJW6
A0A2H1X1H6
A0A1B6H2Y5
+ More
A0A0T6BBI9 A0A2A3E207 A0A1B6CSL3 D6WLY8 A0A1B6HIH9 A0A088ABB8 A0A154P8E5 A0A1S3SJB4 A0A1S3SJC0 A0A1S3SJC6 A0A060X2K8 A0A3P8YQH4 A0A3P8UF30 E0VY35 A0A1S3FS45 F6XFX9 U3CCP8 G1S067 F7EX85 H2PLG7 A0A3B3S576 A0A2J8IW88 H2QU64 A0A087Y613 A0A0L0HHY6 B3KSE9 A0A1U7UYU0 A0A096N6J2 U3IAP7 B2RC85 G2HGX3 A0A3P9IHI0 A0A315V587 A0A2U3VHI9 P0C881 A0A1A8AIS5 A0A2T7NNS4 A0A2I3RKS4 A0A2Y9DUY8 F1RFN1 A0A3B4DK77 A0A340WJM3 A0A1A8MT48 A0A2Y9J711 A0A3Q0HEI0 A0A2K5MJ77 F7EX75 M3YYQ8 A0A226PT86 A0A1V4KS45 A0A3P8PYU0 B3S2M3 A0A369SMW2 A0A1S3J1Z2 A0A1A7WK17 A0A2I3GVU4 A0A1A7Z6D2 C3Y185 A0A2C9JTZ5 A0A224X5J0 A0A2K6GCG5 R7UYD4 A0A1A7XEB6 A0A210Q4K1 A0A2I0M0W1 A0A2I3LIJ4 B2RC85-2 E2R785 A0A267DPT4 A0A2K5BXB7 A0A151N1W9 A0A2J8IWA1 G3T8G9 G3WMG1 A0A3B3YEI5 G1TGH9 A0A2U3YTW7 G3WMG0 A0A2I3S1C3 A0A2U4AZJ2 A0A2U4AZ77 A0A2U4AZ57 H0YFY2 A0A1I8JBR1 A0A2Y9GDP3 G1LR06
A0A0T6BBI9 A0A2A3E207 A0A1B6CSL3 D6WLY8 A0A1B6HIH9 A0A088ABB8 A0A154P8E5 A0A1S3SJB4 A0A1S3SJC0 A0A1S3SJC6 A0A060X2K8 A0A3P8YQH4 A0A3P8UF30 E0VY35 A0A1S3FS45 F6XFX9 U3CCP8 G1S067 F7EX85 H2PLG7 A0A3B3S576 A0A2J8IW88 H2QU64 A0A087Y613 A0A0L0HHY6 B3KSE9 A0A1U7UYU0 A0A096N6J2 U3IAP7 B2RC85 G2HGX3 A0A3P9IHI0 A0A315V587 A0A2U3VHI9 P0C881 A0A1A8AIS5 A0A2T7NNS4 A0A2I3RKS4 A0A2Y9DUY8 F1RFN1 A0A3B4DK77 A0A340WJM3 A0A1A8MT48 A0A2Y9J711 A0A3Q0HEI0 A0A2K5MJ77 F7EX75 M3YYQ8 A0A226PT86 A0A1V4KS45 A0A3P8PYU0 B3S2M3 A0A369SMW2 A0A1S3J1Z2 A0A1A7WK17 A0A2I3GVU4 A0A1A7Z6D2 C3Y185 A0A2C9JTZ5 A0A224X5J0 A0A2K6GCG5 R7UYD4 A0A1A7XEB6 A0A210Q4K1 A0A2I0M0W1 A0A2I3LIJ4 B2RC85-2 E2R785 A0A267DPT4 A0A2K5BXB7 A0A151N1W9 A0A2J8IWA1 G3T8G9 G3WMG1 A0A3B3YEI5 G1TGH9 A0A2U3YTW7 G3WMG0 A0A2I3S1C3 A0A2U4AZJ2 A0A2U4AZ77 A0A2U4AZ57 H0YFY2 A0A1I8JBR1 A0A2Y9GDP3 G1LR06
Pubmed
22118469
26227816
26354079
18362917
19820115
24755649
+ More
25069045 24487278 20566863 25243066 17431167 29240929 16136131 14702039 23749191 12853948 15489334 21484476 17554307 29703783 24621616 18719581 30042472 18563158 15562597 23254933 28812685 23371554 16341006 22293439 21709235 21993624 26392545 20010809
25069045 24487278 20566863 25243066 17431167 29240929 16136131 14702039 23749191 12853948 15489334 21484476 17554307 29703783 24621616 18719581 30042472 18563158 15562597 23254933 28812685 23371554 16341006 22293439 21709235 21993624 26392545 20010809
EMBL
AGBW02009105
OWR51679.1
JTDY01000815
KOB75777.1
KQ461196
KPJ06483.1
+ More
KQ459601 KPI93726.1 ODYU01012724 SOQ59171.1 GECZ01000715 JAS69054.1 LJIG01002246 KRT84694.1 KZ288469 PBC25362.1 GEDC01020973 JAS16325.1 KQ971343 EFA03382.1 GECU01033193 JAS74513.1 KQ434844 KZC08199.1 FR904906 CDQ73487.1 DS235843 EEB18291.1 GAMS01006449 JAB16687.1 ADFV01104281 ADFV01104282 ADFV01104283 ADFV01104284 ADFV01104285 JSUE03027731 JSUE03027732 ABGA01163993 ABGA01163994 ABGA01163995 ABGA01163996 NBAG03000572 PNI14781.1 PNI14782.1 PNI14784.1 AC184156 AC192442 AYCK01000711 KQ257455 KND00707.1 AK093411 BAG52711.1 ADON01076476 AK314983 AC073343 BC034495 BC044242 BC157864 BC157871 AK305987 BAK62981.1 NHOQ01002872 PWA14075.1 AC005995 AA382672 AW474289 HADY01016098 HAEJ01017811 SBP54583.1 PZQS01000010 PVD22818.1 AC193266 AEMK02000014 HAEF01018673 SBR59832.1 AEYP01085086 AEYP01085087 AEYP01085088 AEYP01085089 AEYP01085090 AEYP01085091 AWGT02000022 OXB82710.1 LSYS01001700 OPJ87251.1 DS985247 EDV23121.1 NOWV01000001 RDD47751.1 HADW01004531 SBP05931.1 HADX01015882 SBP38114.1 GG666480 EEN65625.1 GFTR01008669 JAW07757.1 AMQN01005752 KB296765 ELU11324.1 HADW01014938 SBP16338.1 NEDP02005036 OWF43641.1 AKCR02000052 PKK23312.1 AAEX03004285 NIVC01003490 PAA51176.1 AKHW03004154 KYO30645.1 PNI14785.1 AEFK01075606 AEFK01075607 AEFK01075608 AEFK01075609 AEFK01075610 AC079882 ACTA01002360 ACTA01010360 ACTA01018360 ACTA01186349 ACTA01194347
KQ459601 KPI93726.1 ODYU01012724 SOQ59171.1 GECZ01000715 JAS69054.1 LJIG01002246 KRT84694.1 KZ288469 PBC25362.1 GEDC01020973 JAS16325.1 KQ971343 EFA03382.1 GECU01033193 JAS74513.1 KQ434844 KZC08199.1 FR904906 CDQ73487.1 DS235843 EEB18291.1 GAMS01006449 JAB16687.1 ADFV01104281 ADFV01104282 ADFV01104283 ADFV01104284 ADFV01104285 JSUE03027731 JSUE03027732 ABGA01163993 ABGA01163994 ABGA01163995 ABGA01163996 NBAG03000572 PNI14781.1 PNI14782.1 PNI14784.1 AC184156 AC192442 AYCK01000711 KQ257455 KND00707.1 AK093411 BAG52711.1 ADON01076476 AK314983 AC073343 BC034495 BC044242 BC157864 BC157871 AK305987 BAK62981.1 NHOQ01002872 PWA14075.1 AC005995 AA382672 AW474289 HADY01016098 HAEJ01017811 SBP54583.1 PZQS01000010 PVD22818.1 AC193266 AEMK02000014 HAEF01018673 SBR59832.1 AEYP01085086 AEYP01085087 AEYP01085088 AEYP01085089 AEYP01085090 AEYP01085091 AWGT02000022 OXB82710.1 LSYS01001700 OPJ87251.1 DS985247 EDV23121.1 NOWV01000001 RDD47751.1 HADW01004531 SBP05931.1 HADX01015882 SBP38114.1 GG666480 EEN65625.1 GFTR01008669 JAW07757.1 AMQN01005752 KB296765 ELU11324.1 HADW01014938 SBP16338.1 NEDP02005036 OWF43641.1 AKCR02000052 PKK23312.1 AAEX03004285 NIVC01003490 PAA51176.1 AKHW03004154 KYO30645.1 PNI14785.1 AEFK01075606 AEFK01075607 AEFK01075608 AEFK01075609 AEFK01075610 AC079882 ACTA01002360 ACTA01010360 ACTA01018360 ACTA01186349 ACTA01194347
Proteomes
UP000007151
UP000037510
UP000053240
UP000053268
UP000242457
UP000007266
+ More
UP000005203 UP000076502 UP000087266 UP000193380 UP000265140 UP000265120 UP000009046 UP000081671 UP000008225 UP000001073 UP000006718 UP000001595 UP000261540 UP000002277 UP000028760 UP000053201 UP000189704 UP000028761 UP000016666 UP000005640 UP000265200 UP000245340 UP000245119 UP000248480 UP000008227 UP000261440 UP000265300 UP000248482 UP000189705 UP000233060 UP000000715 UP000198419 UP000190648 UP000265100 UP000009022 UP000253843 UP000085678 UP000001554 UP000076420 UP000233160 UP000014760 UP000242188 UP000053872 UP000002254 UP000215902 UP000233020 UP000050525 UP000007646 UP000007648 UP000261480 UP000001811 UP000245341 UP000245320 UP000095280 UP000248481 UP000008912
UP000005203 UP000076502 UP000087266 UP000193380 UP000265140 UP000265120 UP000009046 UP000081671 UP000008225 UP000001073 UP000006718 UP000001595 UP000261540 UP000002277 UP000028760 UP000053201 UP000189704 UP000028761 UP000016666 UP000005640 UP000265200 UP000245340 UP000245119 UP000248480 UP000008227 UP000261440 UP000265300 UP000248482 UP000189705 UP000233060 UP000000715 UP000198419 UP000190648 UP000265100 UP000009022 UP000253843 UP000085678 UP000001554 UP000076420 UP000233160 UP000014760 UP000242188 UP000053872 UP000002254 UP000215902 UP000233020 UP000050525 UP000007646 UP000007648 UP000261480 UP000001811 UP000245341 UP000245320 UP000095280 UP000248481 UP000008912
Interpro
IPR003409
MORN
+ More
IPR042121 MutL_C_regsub
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR002099 DNA_mismatch_repair_N
IPR014762 DNA_mismatch_repair_CS
IPR013507 DNA_mismatch_S5_2-like
IPR037198 MutL_C_sf
IPR036890 HATPase_C_sf
IPR042120 MutL_C_dimsub
IPR014790 MutL_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR003594 HATPase_C
IPR042121 MutL_C_regsub
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR002099 DNA_mismatch_repair_N
IPR014762 DNA_mismatch_repair_CS
IPR013507 DNA_mismatch_S5_2-like
IPR037198 MutL_C_sf
IPR036890 HATPase_C_sf
IPR042120 MutL_C_dimsub
IPR014790 MutL_C
IPR020568 Ribosomal_S5_D2-typ_fold
IPR003594 HATPase_C
ProteinModelPortal
A0A212FD60
A0A0L7LK03
A0A194QLQ1
A0A194PJW6
A0A2H1X1H6
A0A1B6H2Y5
+ More
A0A0T6BBI9 A0A2A3E207 A0A1B6CSL3 D6WLY8 A0A1B6HIH9 A0A088ABB8 A0A154P8E5 A0A1S3SJB4 A0A1S3SJC0 A0A1S3SJC6 A0A060X2K8 A0A3P8YQH4 A0A3P8UF30 E0VY35 A0A1S3FS45 F6XFX9 U3CCP8 G1S067 F7EX85 H2PLG7 A0A3B3S576 A0A2J8IW88 H2QU64 A0A087Y613 A0A0L0HHY6 B3KSE9 A0A1U7UYU0 A0A096N6J2 U3IAP7 B2RC85 G2HGX3 A0A3P9IHI0 A0A315V587 A0A2U3VHI9 P0C881 A0A1A8AIS5 A0A2T7NNS4 A0A2I3RKS4 A0A2Y9DUY8 F1RFN1 A0A3B4DK77 A0A340WJM3 A0A1A8MT48 A0A2Y9J711 A0A3Q0HEI0 A0A2K5MJ77 F7EX75 M3YYQ8 A0A226PT86 A0A1V4KS45 A0A3P8PYU0 B3S2M3 A0A369SMW2 A0A1S3J1Z2 A0A1A7WK17 A0A2I3GVU4 A0A1A7Z6D2 C3Y185 A0A2C9JTZ5 A0A224X5J0 A0A2K6GCG5 R7UYD4 A0A1A7XEB6 A0A210Q4K1 A0A2I0M0W1 A0A2I3LIJ4 B2RC85-2 E2R785 A0A267DPT4 A0A2K5BXB7 A0A151N1W9 A0A2J8IWA1 G3T8G9 G3WMG1 A0A3B3YEI5 G1TGH9 A0A2U3YTW7 G3WMG0 A0A2I3S1C3 A0A2U4AZJ2 A0A2U4AZ77 A0A2U4AZ57 H0YFY2 A0A1I8JBR1 A0A2Y9GDP3 G1LR06
A0A0T6BBI9 A0A2A3E207 A0A1B6CSL3 D6WLY8 A0A1B6HIH9 A0A088ABB8 A0A154P8E5 A0A1S3SJB4 A0A1S3SJC0 A0A1S3SJC6 A0A060X2K8 A0A3P8YQH4 A0A3P8UF30 E0VY35 A0A1S3FS45 F6XFX9 U3CCP8 G1S067 F7EX85 H2PLG7 A0A3B3S576 A0A2J8IW88 H2QU64 A0A087Y613 A0A0L0HHY6 B3KSE9 A0A1U7UYU0 A0A096N6J2 U3IAP7 B2RC85 G2HGX3 A0A3P9IHI0 A0A315V587 A0A2U3VHI9 P0C881 A0A1A8AIS5 A0A2T7NNS4 A0A2I3RKS4 A0A2Y9DUY8 F1RFN1 A0A3B4DK77 A0A340WJM3 A0A1A8MT48 A0A2Y9J711 A0A3Q0HEI0 A0A2K5MJ77 F7EX75 M3YYQ8 A0A226PT86 A0A1V4KS45 A0A3P8PYU0 B3S2M3 A0A369SMW2 A0A1S3J1Z2 A0A1A7WK17 A0A2I3GVU4 A0A1A7Z6D2 C3Y185 A0A2C9JTZ5 A0A224X5J0 A0A2K6GCG5 R7UYD4 A0A1A7XEB6 A0A210Q4K1 A0A2I0M0W1 A0A2I3LIJ4 B2RC85-2 E2R785 A0A267DPT4 A0A2K5BXB7 A0A151N1W9 A0A2J8IWA1 G3T8G9 G3WMG1 A0A3B3YEI5 G1TGH9 A0A2U3YTW7 G3WMG0 A0A2I3S1C3 A0A2U4AZJ2 A0A2U4AZ77 A0A2U4AZ57 H0YFY2 A0A1I8JBR1 A0A2Y9GDP3 G1LR06
Ontologies
KEGG
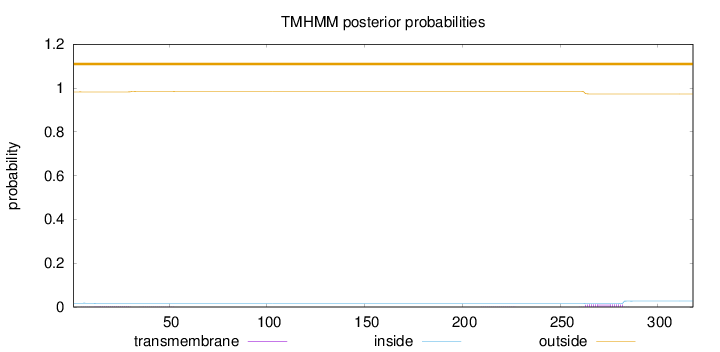
Topology
Length:
318
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.28292
Exp number, first 60 AAs:
0.04607
Total prob of N-in:
0.01767
outside
1 - 318
Population Genetic Test Statistics
Pi
284.889019
Theta
236.327475
Tajima's D
0.784775
CLR
1.311731
CSRT
0.601669916504175
Interpretation
Uncertain
