Gene
KWMTBOMO02071
Pre Gene Modal
BGIBMGA002977
Annotation
PREDICTED:_CDK5_and_ABL1_enzyme_substrate_2_isoform_X2_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.281 Nuclear Reliability : 1.585
Sequence
CDS
ATGGCTGGTATTAATACTGAAGAGAAAAGTAGTTCAGAGAGTCTTACATTTGGTCGCTTCCGTACCAGCAGTACATGTATACCTGAGAACCATATTATGATCAATGAAGTACGTTTTGTAAGATCAACGAAAGGAGTTACATTTAGAGATGAGCGGCTTGCATTTGTGCCGGAAAAAAGTGTACCATGTGCCATTTTCAGCACCATTCCATATTCAAGAACTACAAGAACTGCCAGATCTGAGCTTAGAAAGGATGGAGTGCGCCGAAGAAATACATCAGGACCTAGACCCCTATCTGCAATAACAGACAATGGGGTTGATCCATTTCATCTCTTAGGCTTGGAAAAAGAAGAGAATGGTCAAGACATATCTTATTCTAAACTTCTTGTACCATCTAGAGTACACACATTAGAGCAATACCGTAAATTGTATGATAATGAGTACATTGATAAAAATTGGAAGATATTAAATCGTCACCCTCATGTTATAGCCAGGTGTTTCTCGTACGAGTCTTCCCAAAGGACAACGATAGCGTCCTCACCGCCCCACACTTCTGTTGATAATAAAACGTTCGACTGGGACGAAGGAACTTTGCTTTCACAAAAATATGTTCACTACTGCCCTAATGTACTTGATGATCCAGAACTGATTGCGGGGAAACACAGGACTCTGTTAACGTTTACGTCTTACATGACGTCGATCATTGACTATGTTCGACCGCAGGACTTAAAGAAAGAACTGAATGATAAATTCCGAGAAAAATTCCCACACATTAAACTGACTTTGAGTAAACTAAGAAGTATAAAAAGAGAAATGCGGAAAATCGCCAAACACGACAGCGGAGGGATCGATTTGTTGTCGGTCGCTCAGGCGTACGTTTATTTTGAAAAGTTGATACTCGCGAATCTTATAAACAAGGACAACAGGAAATTATGTGCCGGAGCGTGCCTGTTACTGTCAGCTAAACTAAACGACGTTAAAGGAGAAATTCTGAAGGCGTTGATAGAACGCATAGAGACGACATTCAGACTGGACAGGAAGGATCTGATGAAGTTCGAATTCGCCGTTCTAGTGGCCCTCGAGTTTGGTTTGCACGTGCCGCCGTACGAAGTGTTCCCGCACTACCAGCGCTTGCTCCACGACTCGTGA
Protein
MAGINTEEKSSSESLTFGRFRTSSTCIPENHIMINEVRFVRSTKGVTFRDERLAFVPEKSVPCAIFSTIPYSRTTRTARSELRKDGVRRRNTSGPRPLSAITDNGVDPFHLLGLEKEENGQDISYSKLLVPSRVHTLEQYRKLYDNEYIDKNWKILNRHPHVIARCFSYESSQRTTIASSPPHTSVDNKTFDWDEGTLLSQKYVHYCPNVLDDPELIAGKHRTLLTFTSYMTSIIDYVRPQDLKKELNDKFREKFPHIKLTLSKLRSIKREMRKIAKHDSGGIDLLSVAQAYVYFEKLILANLINKDNRKLCAGACLLLSAKLNDVKGEILKALIERIETTFRLDRKDLMKFEFAVLVALEFGLHVPPYEVFPHYQRLLHDS
Summary
Similarity
Belongs to the cyclin family.
Uniprot
H9J0E1
A0A2A4K6V8
A0A194PK79
A0A212FD78
A0A194QLT4
A0A2J7PBK5
+ More
E0W277 A0A067RGJ6 A0A1B6EDE3 V9IJH3 E2ALX6 A0A0C9PUX4 A0A1S3DKH9 A0A1S3DKW1 A0A195FAA1 A0A088AH88 A0A195BWH5 A0A195CQL7 A0A195ELP7 A0A154P8X6 A0A0N0BG11 A0A026WK63 A0A2A3ES62 A0A0L7QWH7 A0A0K8TMP4 A0A0C9Q588 A0A151X5U3 A0A158NCJ9 F4WX63 A0A0L0C7P7 A0A1I8P2N8 A0A232EG38 A0A3Q0J0C0 A0A1B0D4B0 T1PIY1 E2BM62 A0A1I8MBS8 A0A1I8P2I6 A0A1B6LLG1 A0A1B6I013 A0A1A9XHC0 A0A1A9ZPJ8 Q178L9 A0A1S4FBT2 A0A182GPV5 A0A023EV53 A0A1L8DWW7 A0A1L8DWS0 A0A0K8WGY4 B4GH58 A0A034VD21 W8BAB0 A0A1I8MBS1 A0A1L8DWU3 A0A1B0B813 A0A0A1WKN4 A0A0M4EKY1 V5GWR2 A0A3B0J072 A0A0K8W8J1 A0A034VEL8 B4HQL4 B3NRI7 A0A1W4V4W1 B4QEG2 Q28XX0 A1Z9G1 B4J7A7 A0A0J9U147 W8AMM2 A0A0B4KF19 A0A0A1X6F6 V5GSV0 B4MSA2 A0A3B0J0L3 B4KN84 D6WLY6 B4P481 A0A1W4VH81 A0A0B4KEN8 A0A2S2NVT2 J9JP51 A0A2H8TK10 B3MHD1 A0A1Y1MPZ7 A0A1B0G5D1 N6T4Z0 U4UGW9 A0A2S2PX65 A0A1B0CIB3 B4LMU4 T1J729 A0A336LUY1 V5GNJ8 A0A131Y2Z7 A0A2M4BGR8 A0A2M4BGX5 A0A2M3Z9V6
E0W277 A0A067RGJ6 A0A1B6EDE3 V9IJH3 E2ALX6 A0A0C9PUX4 A0A1S3DKH9 A0A1S3DKW1 A0A195FAA1 A0A088AH88 A0A195BWH5 A0A195CQL7 A0A195ELP7 A0A154P8X6 A0A0N0BG11 A0A026WK63 A0A2A3ES62 A0A0L7QWH7 A0A0K8TMP4 A0A0C9Q588 A0A151X5U3 A0A158NCJ9 F4WX63 A0A0L0C7P7 A0A1I8P2N8 A0A232EG38 A0A3Q0J0C0 A0A1B0D4B0 T1PIY1 E2BM62 A0A1I8MBS8 A0A1I8P2I6 A0A1B6LLG1 A0A1B6I013 A0A1A9XHC0 A0A1A9ZPJ8 Q178L9 A0A1S4FBT2 A0A182GPV5 A0A023EV53 A0A1L8DWW7 A0A1L8DWS0 A0A0K8WGY4 B4GH58 A0A034VD21 W8BAB0 A0A1I8MBS1 A0A1L8DWU3 A0A1B0B813 A0A0A1WKN4 A0A0M4EKY1 V5GWR2 A0A3B0J072 A0A0K8W8J1 A0A034VEL8 B4HQL4 B3NRI7 A0A1W4V4W1 B4QEG2 Q28XX0 A1Z9G1 B4J7A7 A0A0J9U147 W8AMM2 A0A0B4KF19 A0A0A1X6F6 V5GSV0 B4MSA2 A0A3B0J0L3 B4KN84 D6WLY6 B4P481 A0A1W4VH81 A0A0B4KEN8 A0A2S2NVT2 J9JP51 A0A2H8TK10 B3MHD1 A0A1Y1MPZ7 A0A1B0G5D1 N6T4Z0 U4UGW9 A0A2S2PX65 A0A1B0CIB3 B4LMU4 T1J729 A0A336LUY1 V5GNJ8 A0A131Y2Z7 A0A2M4BGR8 A0A2M4BGX5 A0A2M3Z9V6
Pubmed
19121390
26354079
22118469
20566863
24845553
20798317
+ More
24508170 30249741 26369729 21347285 21719571 26108605 28648823 25315136 17510324 26483478 24945155 17994087 25348373 24495485 25830018 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 17550304 28004739 23537049 25765539
24508170 30249741 26369729 21347285 21719571 26108605 28648823 25315136 17510324 26483478 24945155 17994087 25348373 24495485 25830018 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 18362917 19820115 17550304 28004739 23537049 25765539
EMBL
BABH01010521
BABH01010522
NWSH01000081
PCG79809.1
KQ459601
KPI93727.1
+ More
AGBW02009105 OWR51680.1 KQ461196 KPJ06487.1 NEVH01027088 PNF13702.1 DS235874 EEB19733.1 KK852482 KDR22907.1 GEDC01001414 JAS35884.1 JR050440 AEY61278.1 GL440682 EFN65568.1 GBYB01005218 JAG74985.1 KQ981727 KYN36974.1 KQ976396 KYM92984.1 KQ977408 KYN02940.1 KQ978691 KYN29200.1 KQ434827 KZC07600.1 KQ435794 KOX73891.1 KK107167 QOIP01000010 EZA56363.1 RLU18052.1 KZ288189 PBC34633.1 KQ414713 KOC62967.1 GDAI01001986 JAI15617.1 GBYB01009218 JAG78985.1 KQ982490 KYQ55783.1 ADTU01011950 ADTU01011951 GL888417 EGI61306.1 JRES01000803 KNC28282.1 NNAY01004878 OXU17307.1 AJVK01011336 AJVK01011337 KA648070 AFP62699.1 GL449158 EFN83188.1 GEBQ01015434 JAT24543.1 GECU01028238 GECU01027456 JAS79468.1 JAS80250.1 CH477362 EAT42623.1 JXUM01016532 JXUM01016533 KQ560461 KXJ82300.1 GAPW01000727 JAC12871.1 GFDF01003210 JAV10874.1 GFDF01003211 JAV10873.1 GDHF01024349 GDHF01016422 GDHF01007288 GDHF01002005 JAI27965.1 JAI35892.1 JAI45026.1 JAI50309.1 CH479183 EDW35828.1 GAKP01018935 JAC40017.1 GAMC01016544 JAB90011.1 GFDF01003212 JAV10872.1 JXJN01009799 JXJN01009800 GBXI01015207 JAC99084.1 CP012524 ALC42130.1 GALX01003763 JAB64703.1 OUUW01000001 SPP74244.1 GDHF01029306 GDHF01011346 GDHF01004975 JAI23008.1 JAI40968.1 JAI47339.1 GAKP01018934 JAC40018.1 CH480816 EDW47749.1 CH954179 EDV56139.2 CM000362 EDX06952.1 CM000071 EAL26196.3 AE013599 BT133388 AAF58349.1 AFH41847.1 CH916367 EDW02125.1 CM002911 KMY93545.1 GAMC01016545 JAB90010.1 AGB93490.1 GBXI01007811 GBXI01001955 JAD06481.1 JAD12337.1 GALX01003764 JAB64702.1 CH963850 EDW74991.2 SPP74245.1 CH933808 EDW09937.2 KQ971343 EFA03383.2 CM000158 EDW90591.2 AGB93489.1 GGMR01008704 MBY21323.1 ABLF02020824 ABLF02020825 GFXV01001813 MBW13618.1 CH902619 EDV37931.2 GEZM01029634 JAV86026.1 CCAG010020791 CCAG010020792 APGK01043870 KB741018 ENN75229.1 KB632152 ERL89140.1 GGMS01000751 MBY69954.1 AJWK01013177 AJWK01013178 AJWK01013179 CH940648 EDW61035.2 JH431907 UFQT01000124 SSX20483.1 GANP01012548 JAB71920.1 GEFM01002944 JAP72852.1 GGFJ01003114 MBW52255.1 GGFJ01003165 MBW52306.1 GGFM01004550 MBW25301.1
AGBW02009105 OWR51680.1 KQ461196 KPJ06487.1 NEVH01027088 PNF13702.1 DS235874 EEB19733.1 KK852482 KDR22907.1 GEDC01001414 JAS35884.1 JR050440 AEY61278.1 GL440682 EFN65568.1 GBYB01005218 JAG74985.1 KQ981727 KYN36974.1 KQ976396 KYM92984.1 KQ977408 KYN02940.1 KQ978691 KYN29200.1 KQ434827 KZC07600.1 KQ435794 KOX73891.1 KK107167 QOIP01000010 EZA56363.1 RLU18052.1 KZ288189 PBC34633.1 KQ414713 KOC62967.1 GDAI01001986 JAI15617.1 GBYB01009218 JAG78985.1 KQ982490 KYQ55783.1 ADTU01011950 ADTU01011951 GL888417 EGI61306.1 JRES01000803 KNC28282.1 NNAY01004878 OXU17307.1 AJVK01011336 AJVK01011337 KA648070 AFP62699.1 GL449158 EFN83188.1 GEBQ01015434 JAT24543.1 GECU01028238 GECU01027456 JAS79468.1 JAS80250.1 CH477362 EAT42623.1 JXUM01016532 JXUM01016533 KQ560461 KXJ82300.1 GAPW01000727 JAC12871.1 GFDF01003210 JAV10874.1 GFDF01003211 JAV10873.1 GDHF01024349 GDHF01016422 GDHF01007288 GDHF01002005 JAI27965.1 JAI35892.1 JAI45026.1 JAI50309.1 CH479183 EDW35828.1 GAKP01018935 JAC40017.1 GAMC01016544 JAB90011.1 GFDF01003212 JAV10872.1 JXJN01009799 JXJN01009800 GBXI01015207 JAC99084.1 CP012524 ALC42130.1 GALX01003763 JAB64703.1 OUUW01000001 SPP74244.1 GDHF01029306 GDHF01011346 GDHF01004975 JAI23008.1 JAI40968.1 JAI47339.1 GAKP01018934 JAC40018.1 CH480816 EDW47749.1 CH954179 EDV56139.2 CM000362 EDX06952.1 CM000071 EAL26196.3 AE013599 BT133388 AAF58349.1 AFH41847.1 CH916367 EDW02125.1 CM002911 KMY93545.1 GAMC01016545 JAB90010.1 AGB93490.1 GBXI01007811 GBXI01001955 JAD06481.1 JAD12337.1 GALX01003764 JAB64702.1 CH963850 EDW74991.2 SPP74245.1 CH933808 EDW09937.2 KQ971343 EFA03383.2 CM000158 EDW90591.2 AGB93489.1 GGMR01008704 MBY21323.1 ABLF02020824 ABLF02020825 GFXV01001813 MBW13618.1 CH902619 EDV37931.2 GEZM01029634 JAV86026.1 CCAG010020791 CCAG010020792 APGK01043870 KB741018 ENN75229.1 KB632152 ERL89140.1 GGMS01000751 MBY69954.1 AJWK01013177 AJWK01013178 AJWK01013179 CH940648 EDW61035.2 JH431907 UFQT01000124 SSX20483.1 GANP01012548 JAB71920.1 GEFM01002944 JAP72852.1 GGFJ01003114 MBW52255.1 GGFJ01003165 MBW52306.1 GGFM01004550 MBW25301.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000235965
+ More
UP000009046 UP000027135 UP000000311 UP000079169 UP000078541 UP000005203 UP000078540 UP000078542 UP000078492 UP000076502 UP000053105 UP000053097 UP000279307 UP000242457 UP000053825 UP000075809 UP000005205 UP000007755 UP000037069 UP000095300 UP000215335 UP000092462 UP000008237 UP000095301 UP000092443 UP000092445 UP000008820 UP000069940 UP000249989 UP000008744 UP000092460 UP000092553 UP000268350 UP000001292 UP000008711 UP000192221 UP000000304 UP000001819 UP000000803 UP000001070 UP000007798 UP000009192 UP000007266 UP000002282 UP000007819 UP000007801 UP000092444 UP000019118 UP000030742 UP000092461 UP000008792
UP000009046 UP000027135 UP000000311 UP000079169 UP000078541 UP000005203 UP000078540 UP000078542 UP000078492 UP000076502 UP000053105 UP000053097 UP000279307 UP000242457 UP000053825 UP000075809 UP000005205 UP000007755 UP000037069 UP000095300 UP000215335 UP000092462 UP000008237 UP000095301 UP000092443 UP000092445 UP000008820 UP000069940 UP000249989 UP000008744 UP000092460 UP000092553 UP000268350 UP000001292 UP000008711 UP000192221 UP000000304 UP000001819 UP000000803 UP000001070 UP000007798 UP000009192 UP000007266 UP000002282 UP000007819 UP000007801 UP000092444 UP000019118 UP000030742 UP000092461 UP000008792
Pfam
PF00134 Cyclin_N
Interpro
SUPFAM
SSF47954
SSF47954
CDD
ProteinModelPortal
H9J0E1
A0A2A4K6V8
A0A194PK79
A0A212FD78
A0A194QLT4
A0A2J7PBK5
+ More
E0W277 A0A067RGJ6 A0A1B6EDE3 V9IJH3 E2ALX6 A0A0C9PUX4 A0A1S3DKH9 A0A1S3DKW1 A0A195FAA1 A0A088AH88 A0A195BWH5 A0A195CQL7 A0A195ELP7 A0A154P8X6 A0A0N0BG11 A0A026WK63 A0A2A3ES62 A0A0L7QWH7 A0A0K8TMP4 A0A0C9Q588 A0A151X5U3 A0A158NCJ9 F4WX63 A0A0L0C7P7 A0A1I8P2N8 A0A232EG38 A0A3Q0J0C0 A0A1B0D4B0 T1PIY1 E2BM62 A0A1I8MBS8 A0A1I8P2I6 A0A1B6LLG1 A0A1B6I013 A0A1A9XHC0 A0A1A9ZPJ8 Q178L9 A0A1S4FBT2 A0A182GPV5 A0A023EV53 A0A1L8DWW7 A0A1L8DWS0 A0A0K8WGY4 B4GH58 A0A034VD21 W8BAB0 A0A1I8MBS1 A0A1L8DWU3 A0A1B0B813 A0A0A1WKN4 A0A0M4EKY1 V5GWR2 A0A3B0J072 A0A0K8W8J1 A0A034VEL8 B4HQL4 B3NRI7 A0A1W4V4W1 B4QEG2 Q28XX0 A1Z9G1 B4J7A7 A0A0J9U147 W8AMM2 A0A0B4KF19 A0A0A1X6F6 V5GSV0 B4MSA2 A0A3B0J0L3 B4KN84 D6WLY6 B4P481 A0A1W4VH81 A0A0B4KEN8 A0A2S2NVT2 J9JP51 A0A2H8TK10 B3MHD1 A0A1Y1MPZ7 A0A1B0G5D1 N6T4Z0 U4UGW9 A0A2S2PX65 A0A1B0CIB3 B4LMU4 T1J729 A0A336LUY1 V5GNJ8 A0A131Y2Z7 A0A2M4BGR8 A0A2M4BGX5 A0A2M3Z9V6
E0W277 A0A067RGJ6 A0A1B6EDE3 V9IJH3 E2ALX6 A0A0C9PUX4 A0A1S3DKH9 A0A1S3DKW1 A0A195FAA1 A0A088AH88 A0A195BWH5 A0A195CQL7 A0A195ELP7 A0A154P8X6 A0A0N0BG11 A0A026WK63 A0A2A3ES62 A0A0L7QWH7 A0A0K8TMP4 A0A0C9Q588 A0A151X5U3 A0A158NCJ9 F4WX63 A0A0L0C7P7 A0A1I8P2N8 A0A232EG38 A0A3Q0J0C0 A0A1B0D4B0 T1PIY1 E2BM62 A0A1I8MBS8 A0A1I8P2I6 A0A1B6LLG1 A0A1B6I013 A0A1A9XHC0 A0A1A9ZPJ8 Q178L9 A0A1S4FBT2 A0A182GPV5 A0A023EV53 A0A1L8DWW7 A0A1L8DWS0 A0A0K8WGY4 B4GH58 A0A034VD21 W8BAB0 A0A1I8MBS1 A0A1L8DWU3 A0A1B0B813 A0A0A1WKN4 A0A0M4EKY1 V5GWR2 A0A3B0J072 A0A0K8W8J1 A0A034VEL8 B4HQL4 B3NRI7 A0A1W4V4W1 B4QEG2 Q28XX0 A1Z9G1 B4J7A7 A0A0J9U147 W8AMM2 A0A0B4KF19 A0A0A1X6F6 V5GSV0 B4MSA2 A0A3B0J0L3 B4KN84 D6WLY6 B4P481 A0A1W4VH81 A0A0B4KEN8 A0A2S2NVT2 J9JP51 A0A2H8TK10 B3MHD1 A0A1Y1MPZ7 A0A1B0G5D1 N6T4Z0 U4UGW9 A0A2S2PX65 A0A1B0CIB3 B4LMU4 T1J729 A0A336LUY1 V5GNJ8 A0A131Y2Z7 A0A2M4BGR8 A0A2M4BGX5 A0A2M3Z9V6
Ontologies
PANTHER
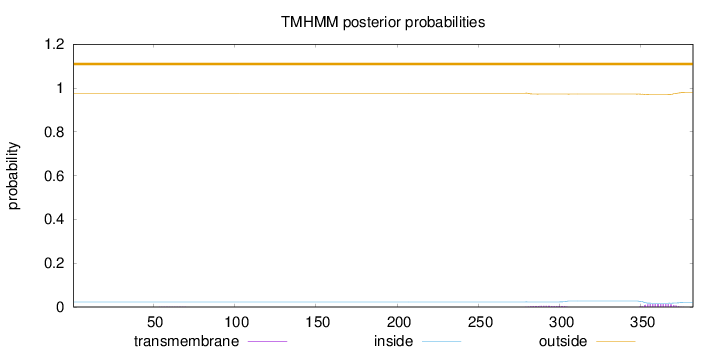
Topology
Length:
382
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.37025
Exp number, first 60 AAs:
0.00101
Total prob of N-in:
0.02332
outside
1 - 382
Population Genetic Test Statistics
Pi
270.28508
Theta
194.465476
Tajima's D
1.222396
CLR
0
CSRT
0.72046397680116
Interpretation
Uncertain
