Gene
KWMTBOMO02070
Pre Gene Modal
BGIBMGA003192
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_MIB1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.011
Sequence
CDS
ATGGAGAGTAATATCTCTGTTTCATCAACATCATCTAGACCAACCCGTTTTATGATGGAAGGAGTAGGAGCTCGTGTGATTCGAGGTCCAGATTGGAAGTGGGGAAAACAGGATGGAGGAGAGGGCCATCTTGGCACTGTAAGGAATTTTGAGTCTCCAGAAGAAGTAGTAGTCGTTTGGGATAATGGAACAGCTGCAAATTATAGATGTTCAGGTGCCTATGATCTGCGGATTCTGGATAGTGCCCCTACTGGTGTGAAACATGAAGGAACTATGTGTGATACTTGCAGGCAACAACCCATATTTGGTATACGTTGGAAATGTGCTGAATGCAGCAATTATGATCTATGCTCTGTCTGCTATCATGGAGATAAACATAATTTACGTCATAGATTTTATAGAATTAGTGCACCAGGGGCACAACGATGTTTGGTTGAACCACGAAGAAAAAGTAAAAAACAGGCTGTCAGGGGTATCTTTCCAGGAGCCAGAGTCGTAAGAGGGGTGGATTGGCAGTGGGAAGATCAAGATGGTGGAAATTGTAGGCGTGGAAAAGTCAATGAAATCCAAGATTGGTCAGCAGCTAGTCCACGATCTGCTGCATATGTTGTGTGGGATAATGGTGCCAAAAATCTCTACAGGGTAGGCTTTGAGGGAATGGCTGACCTGAAGGCCGTTAATGATGTGAAAGGTCAGAATGTGTACAAGGAACACTTGCCATTGCTAGGCGAACTTGGCCCTGGAAGAACAGGCCCACATGGACTACAAGTTGGTGATCAGGTGAATGTAGACCTTGATCTTGAAATAGTACAGTCACTACAACATGGTCATGGTGGTTGGACAGATGGTATGTTCGAGTGTCTCGGCACCACCGGTACTGTGGTTGGCATAGATGAGGACCACGACATTGTTGTAACTTATCCAAGCGGAAATAGGTGGACATTTAACCCTGCTGTTTTAACTAAGATATTCAGCGGCGCGTACACGACGACGAGCCCGACGGGCGGCGCGGGCGCGGCGGGTGCGGGCGGAGGGTTCGCGGTGGGAGACCTCGTGCAGGTCTGCGCCGACCAGGAGCGGGTCAAGGCGCTGCAGCGAGGACACGGGGAGTGGGCCGAGGCCATGGCACCTACTCTTGGCAAGATAGGACGAGTGCAGCAGATTTATCACGACAACGATCTAAAGGTGGAAGTGTGTAATACGTCATGGACGTACAATCCGAACGCGGTGACAAAAGTTGCGTCGTCCGATGGCAGCATTCCGGGAAACTCTTCAGGTGAACGGTTATCGGCGCTTTTAAAAAAGTTATTTGAATCCCACGTGACCGGAGACGCAAACGAGGAGCTGGTGAAGGCGGCCGCCAATGGCGACGCGACGCGCTGCGCCGAGATACTGGCGCGCACCGACGGCGCACAGGCTGACGTCAACGGCTTCTACGGCGGACACACCGCGCTGCAGGCGGCCGCGCAGAACGGCCACGTGGAGGTGATCCGCGCGCTGGTGGAGGCGGGCGCGGAGGTGGACGCGGAGGACCGCGACGGGGACCGCGCCGCGCACCACGCCGCCTTCGGGGACGAGCCGGCCGCCCTGCGCGCGCTGGGCGCCGCCGCCGCCGACCTCAACGCGCGCAACCGCCGCCGCCAGACGCCGCTGCACATCGCCGTCAACAAGGGACACCTCGGCGTCGTGCGCACGCTGCTGCAGCTCGCCGTGCACCCCGCCCTGCAGGACTCTGAAGGCGATACACCCCTGCATGATGCCATTTCAAAAAAGCGTGACGACATGCTAACGCTGCTACTAGAACATGGAGCCGATATGACCCTAACAAACAATAATGGTTTCAACGCTCTGCATCACGCGGCGCTCCGCGGCAATCCAAGGTAA
Protein
MESNISVSSTSSRPTRFMMEGVGARVIRGPDWKWGKQDGGEGHLGTVRNFESPEEVVVVWDNGTAANYRCSGAYDLRILDSAPTGVKHEGTMCDTCRQQPIFGIRWKCAECSNYDLCSVCYHGDKHNLRHRFYRISAPGAQRCLVEPRRKSKKQAVRGIFPGARVVRGVDWQWEDQDGGNCRRGKVNEIQDWSAASPRSAAYVVWDNGAKNLYRVGFEGMADLKAVNDVKGQNVYKEHLPLLGELGPGRTGPHGLQVGDQVNVDLDLEIVQSLQHGHGGWTDGMFECLGTTGTVVGIDEDHDIVVTYPSGNRWTFNPAVLTKIFSGAYTTTSPTGGAGAAGAGGGFAVGDLVQVCADQERVKALQRGHGEWAEAMAPTLGKIGRVQQIYHDNDLKVEVCNTSWTYNPNAVTKVASSDGSIPGNSSGERLSALLKKLFESHVTGDANEELVKAAANGDATRCAEILARTDGAQADVNGFYGGHTALQAAAQNGHVEVIRALVEAGAEVDAEDRDGDRAAHHAAFGDEPAALRALGAAAADLNARNRRRQTPLHIAVNKGHLGVVRTLLQLAVHPALQDSEGDTPLHDAISKKRDDMLTLLLEHGADMTLTNNNGFNALHHAALRGNPR
Summary
Uniprot
A0A2A4K825
A0A2H1WYW2
A0A067QXM4
A0A1B6L2V7
A0A158NK40
A0A3L8DMJ2
+ More
A0A195BIS2 A0A088A2Y4 T1IMQ6 A0A1S3K889 A0A0C9RRG6 A0A1S3IWZ1 A0A195F4F6 A0A1S3K8D5 A0A0P4W1M4 A0A2R5LM51 A0A087T2H4 A0A210QJB6 A0A0N8EAU7 A0A0P4XZ39 A0A0P5LK51 A0A0P5L012 A0A0P5H436 A0A0P6H0L9 A0A165A7T5 A0A0N8C9Q3 A0A0P5JK59 A0A0P5QQ02 A0A0P4XVE7 A0A0P5YD86 A0A0P4ZHZ0 A0A0P5N877 A0A0P6EFF0 A0A0P5IE16 A0A0P5K2I7 A0A2T7NNH6 A0A0P5I4X8 A0A0N8B2C8 K1QEY5 A0A0P5TUW7 A0A0P5I8G9 A0A0P5DUB9 A0A0N8DKE9 A0A0P5R4A7 A0A0P5R7A7 A0A0P5Q991 A0A0P5YTS7 A0A131YG45 A0A0P5D2Z2 A0A0N8CDC2 A0A0P5DGS8 A0A224YXG4 A0A0P5VYU5 A0A0P5UKB3 A0A147BPD1 A0A0P5NJF4 A0A0P5G0C5 A0A0P5XCK9 A0A1Y1KUT7 A0A131XRY8 A0A139WH71 A0A0P5LCI5 A0A1W4X289 A0A0P6DP87 A0A0P5QEQ0 A0A1S4FUL6 Q16NN6 A0A182H367 A0A182PCA1 A0A182JPY1 A0A084WII2 A0A182J119 A0A182FB79 W5JW25 A0A0P5RS99 A0A182QLH9 A0A182Y9I2 A0A2M4A7V1 A0A182N2S7 A0A0U2UAP2 A0A336KUF3 A0A1L8DSE2 A0A182RVG4 A0A182LWW5 A0A182WE12 A0A1Q3FAY6 A0A146LRU8 A0A182HHM1 Q7PZI6 A0A182UTQ6 A0A023F2T6 A0A182L569 A0A0P6HXA4 A0A0P4VNN6 A0A182U438 A0A0A9WIY7 A0A0A1WVB5 A0A0V0G4N4 A0A069DZQ0 A0A1I8MXZ1
A0A195BIS2 A0A088A2Y4 T1IMQ6 A0A1S3K889 A0A0C9RRG6 A0A1S3IWZ1 A0A195F4F6 A0A1S3K8D5 A0A0P4W1M4 A0A2R5LM51 A0A087T2H4 A0A210QJB6 A0A0N8EAU7 A0A0P4XZ39 A0A0P5LK51 A0A0P5L012 A0A0P5H436 A0A0P6H0L9 A0A165A7T5 A0A0N8C9Q3 A0A0P5JK59 A0A0P5QQ02 A0A0P4XVE7 A0A0P5YD86 A0A0P4ZHZ0 A0A0P5N877 A0A0P6EFF0 A0A0P5IE16 A0A0P5K2I7 A0A2T7NNH6 A0A0P5I4X8 A0A0N8B2C8 K1QEY5 A0A0P5TUW7 A0A0P5I8G9 A0A0P5DUB9 A0A0N8DKE9 A0A0P5R4A7 A0A0P5R7A7 A0A0P5Q991 A0A0P5YTS7 A0A131YG45 A0A0P5D2Z2 A0A0N8CDC2 A0A0P5DGS8 A0A224YXG4 A0A0P5VYU5 A0A0P5UKB3 A0A147BPD1 A0A0P5NJF4 A0A0P5G0C5 A0A0P5XCK9 A0A1Y1KUT7 A0A131XRY8 A0A139WH71 A0A0P5LCI5 A0A1W4X289 A0A0P6DP87 A0A0P5QEQ0 A0A1S4FUL6 Q16NN6 A0A182H367 A0A182PCA1 A0A182JPY1 A0A084WII2 A0A182J119 A0A182FB79 W5JW25 A0A0P5RS99 A0A182QLH9 A0A182Y9I2 A0A2M4A7V1 A0A182N2S7 A0A0U2UAP2 A0A336KUF3 A0A1L8DSE2 A0A182RVG4 A0A182LWW5 A0A182WE12 A0A1Q3FAY6 A0A146LRU8 A0A182HHM1 Q7PZI6 A0A182UTQ6 A0A023F2T6 A0A182L569 A0A0P6HXA4 A0A0P4VNN6 A0A182U438 A0A0A9WIY7 A0A0A1WVB5 A0A0V0G4N4 A0A069DZQ0 A0A1I8MXZ1
Pubmed
EMBL
NWSH01000081
PCG79810.1
ODYU01012057
SOQ58156.1
KK852840
KDR15200.1
+ More
GEBQ01021905 JAT18072.1 ADTU01018529 ADTU01018530 ADTU01018531 ADTU01018532 ADTU01018533 ADTU01018534 QOIP01000006 RLU21640.1 KQ976465 KYM84257.1 JH431092 GBYB01009931 JAG79698.1 KQ981820 KYN35266.1 GDRN01079950 JAI62310.1 GGLE01006382 MBY10508.1 KK113087 KFM59313.1 NEDP02003363 OWF48875.1 GDIQ01044892 JAN49845.1 GDIP01238623 JAI84778.1 GDIQ01177145 JAK74580.1 GDIQ01177146 JAK74579.1 GDIQ01231689 GDIQ01072636 JAK20036.1 GDIQ01036547 JAN58190.1 LRGB01000642 KZS17276.1 GDIQ01101035 JAL50691.1 GDIQ01201904 JAK49821.1 GDIQ01121032 GDIQ01080234 GDIQ01030329 JAL30694.1 GDIP01237263 JAI86138.1 GDIP01059388 JAM44327.1 GDIP01216412 JAJ06990.1 GDIQ01157055 JAK94670.1 GDIQ01063440 JAN31297.1 GDIQ01214682 JAK37043.1 GDIQ01216236 JAK35489.1 PZQS01000010 PVD22714.1 GDIQ01218490 JAK33235.1 GDIQ01219635 JAK32090.1 JH817657 EKC29654.1 GDIP01121320 JAL82394.1 GDIQ01217338 JAK34387.1 GDIP01157523 JAJ65879.1 GDIP01025055 JAM78660.1 GDIQ01115226 JAL36500.1 GDIQ01114020 JAL37706.1 GDIQ01138969 JAL12757.1 GDIP01053491 GDIP01053490 JAM50224.1 GEDV01010273 JAP78284.1 GDIP01163669 JAJ59733.1 GDIP01142871 JAL60843.1 GDIP01156479 JAJ66923.1 GFPF01007498 MAA18644.1 GDIP01093278 JAM10437.1 GDIP01111675 JAL92039.1 GEGO01002788 JAR92616.1 GDIQ01151706 JAL00020.1 GDIQ01253080 JAJ98644.1 GDIP01074090 JAM29625.1 GEZM01075200 JAV64211.1 GEFM01006946 JAP68850.1 KQ971343 KYB27259.1 GDIQ01171703 JAK80022.1 GDIQ01089274 JAN05463.1 GDIQ01136519 JAL15207.1 CH477816 EAT35949.1 JXUM01106761 JXUM01106762 JXUM01106763 KQ565082 KXJ71361.1 ATLV01023934 KE525347 KFB50026.1 ADMH02000120 ETN67713.1 GDIQ01115225 JAL36501.1 AXCN02001087 GGFK01003397 MBW36718.1 KT984258 ALR88655.1 UFQS01000833 UFQT01000833 SSX07125.1 SSX27468.1 GFDF01004688 JAV09396.1 AXCM01001233 GFDL01010356 JAV24689.1 GDHC01008081 JAQ10548.1 APCN01002428 AAAB01008986 EAA00278.4 GBBI01003368 JAC15344.1 GDIQ01020721 JAN74016.1 GDKW01000313 JAI56282.1 GBHO01038787 GBHO01038784 JAG04817.1 JAG04820.1 GBXI01011822 JAD02470.1 GECL01003773 JAP02351.1 GBGD01000255 JAC88634.1
GEBQ01021905 JAT18072.1 ADTU01018529 ADTU01018530 ADTU01018531 ADTU01018532 ADTU01018533 ADTU01018534 QOIP01000006 RLU21640.1 KQ976465 KYM84257.1 JH431092 GBYB01009931 JAG79698.1 KQ981820 KYN35266.1 GDRN01079950 JAI62310.1 GGLE01006382 MBY10508.1 KK113087 KFM59313.1 NEDP02003363 OWF48875.1 GDIQ01044892 JAN49845.1 GDIP01238623 JAI84778.1 GDIQ01177145 JAK74580.1 GDIQ01177146 JAK74579.1 GDIQ01231689 GDIQ01072636 JAK20036.1 GDIQ01036547 JAN58190.1 LRGB01000642 KZS17276.1 GDIQ01101035 JAL50691.1 GDIQ01201904 JAK49821.1 GDIQ01121032 GDIQ01080234 GDIQ01030329 JAL30694.1 GDIP01237263 JAI86138.1 GDIP01059388 JAM44327.1 GDIP01216412 JAJ06990.1 GDIQ01157055 JAK94670.1 GDIQ01063440 JAN31297.1 GDIQ01214682 JAK37043.1 GDIQ01216236 JAK35489.1 PZQS01000010 PVD22714.1 GDIQ01218490 JAK33235.1 GDIQ01219635 JAK32090.1 JH817657 EKC29654.1 GDIP01121320 JAL82394.1 GDIQ01217338 JAK34387.1 GDIP01157523 JAJ65879.1 GDIP01025055 JAM78660.1 GDIQ01115226 JAL36500.1 GDIQ01114020 JAL37706.1 GDIQ01138969 JAL12757.1 GDIP01053491 GDIP01053490 JAM50224.1 GEDV01010273 JAP78284.1 GDIP01163669 JAJ59733.1 GDIP01142871 JAL60843.1 GDIP01156479 JAJ66923.1 GFPF01007498 MAA18644.1 GDIP01093278 JAM10437.1 GDIP01111675 JAL92039.1 GEGO01002788 JAR92616.1 GDIQ01151706 JAL00020.1 GDIQ01253080 JAJ98644.1 GDIP01074090 JAM29625.1 GEZM01075200 JAV64211.1 GEFM01006946 JAP68850.1 KQ971343 KYB27259.1 GDIQ01171703 JAK80022.1 GDIQ01089274 JAN05463.1 GDIQ01136519 JAL15207.1 CH477816 EAT35949.1 JXUM01106761 JXUM01106762 JXUM01106763 KQ565082 KXJ71361.1 ATLV01023934 KE525347 KFB50026.1 ADMH02000120 ETN67713.1 GDIQ01115225 JAL36501.1 AXCN02001087 GGFK01003397 MBW36718.1 KT984258 ALR88655.1 UFQS01000833 UFQT01000833 SSX07125.1 SSX27468.1 GFDF01004688 JAV09396.1 AXCM01001233 GFDL01010356 JAV24689.1 GDHC01008081 JAQ10548.1 APCN01002428 AAAB01008986 EAA00278.4 GBBI01003368 JAC15344.1 GDIQ01020721 JAN74016.1 GDKW01000313 JAI56282.1 GBHO01038787 GBHO01038784 JAG04817.1 JAG04820.1 GBXI01011822 JAD02470.1 GECL01003773 JAP02351.1 GBGD01000255 JAC88634.1
Proteomes
UP000218220
UP000027135
UP000005205
UP000279307
UP000078540
UP000005203
+ More
UP000085678 UP000078541 UP000054359 UP000242188 UP000076858 UP000245119 UP000005408 UP000007266 UP000192223 UP000008820 UP000069940 UP000249989 UP000075885 UP000075881 UP000030765 UP000075880 UP000069272 UP000000673 UP000075886 UP000076408 UP000075884 UP000075900 UP000075883 UP000075920 UP000075840 UP000007062 UP000075903 UP000075882 UP000075902 UP000095301
UP000085678 UP000078541 UP000054359 UP000242188 UP000076858 UP000245119 UP000005408 UP000007266 UP000192223 UP000008820 UP000069940 UP000249989 UP000075885 UP000075881 UP000030765 UP000075880 UP000069272 UP000000673 UP000075886 UP000076408 UP000075884 UP000075900 UP000075883 UP000075920 UP000075840 UP000007062 UP000075903 UP000075882 UP000075902 UP000095301
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K825
A0A2H1WYW2
A0A067QXM4
A0A1B6L2V7
A0A158NK40
A0A3L8DMJ2
+ More
A0A195BIS2 A0A088A2Y4 T1IMQ6 A0A1S3K889 A0A0C9RRG6 A0A1S3IWZ1 A0A195F4F6 A0A1S3K8D5 A0A0P4W1M4 A0A2R5LM51 A0A087T2H4 A0A210QJB6 A0A0N8EAU7 A0A0P4XZ39 A0A0P5LK51 A0A0P5L012 A0A0P5H436 A0A0P6H0L9 A0A165A7T5 A0A0N8C9Q3 A0A0P5JK59 A0A0P5QQ02 A0A0P4XVE7 A0A0P5YD86 A0A0P4ZHZ0 A0A0P5N877 A0A0P6EFF0 A0A0P5IE16 A0A0P5K2I7 A0A2T7NNH6 A0A0P5I4X8 A0A0N8B2C8 K1QEY5 A0A0P5TUW7 A0A0P5I8G9 A0A0P5DUB9 A0A0N8DKE9 A0A0P5R4A7 A0A0P5R7A7 A0A0P5Q991 A0A0P5YTS7 A0A131YG45 A0A0P5D2Z2 A0A0N8CDC2 A0A0P5DGS8 A0A224YXG4 A0A0P5VYU5 A0A0P5UKB3 A0A147BPD1 A0A0P5NJF4 A0A0P5G0C5 A0A0P5XCK9 A0A1Y1KUT7 A0A131XRY8 A0A139WH71 A0A0P5LCI5 A0A1W4X289 A0A0P6DP87 A0A0P5QEQ0 A0A1S4FUL6 Q16NN6 A0A182H367 A0A182PCA1 A0A182JPY1 A0A084WII2 A0A182J119 A0A182FB79 W5JW25 A0A0P5RS99 A0A182QLH9 A0A182Y9I2 A0A2M4A7V1 A0A182N2S7 A0A0U2UAP2 A0A336KUF3 A0A1L8DSE2 A0A182RVG4 A0A182LWW5 A0A182WE12 A0A1Q3FAY6 A0A146LRU8 A0A182HHM1 Q7PZI6 A0A182UTQ6 A0A023F2T6 A0A182L569 A0A0P6HXA4 A0A0P4VNN6 A0A182U438 A0A0A9WIY7 A0A0A1WVB5 A0A0V0G4N4 A0A069DZQ0 A0A1I8MXZ1
A0A195BIS2 A0A088A2Y4 T1IMQ6 A0A1S3K889 A0A0C9RRG6 A0A1S3IWZ1 A0A195F4F6 A0A1S3K8D5 A0A0P4W1M4 A0A2R5LM51 A0A087T2H4 A0A210QJB6 A0A0N8EAU7 A0A0P4XZ39 A0A0P5LK51 A0A0P5L012 A0A0P5H436 A0A0P6H0L9 A0A165A7T5 A0A0N8C9Q3 A0A0P5JK59 A0A0P5QQ02 A0A0P4XVE7 A0A0P5YD86 A0A0P4ZHZ0 A0A0P5N877 A0A0P6EFF0 A0A0P5IE16 A0A0P5K2I7 A0A2T7NNH6 A0A0P5I4X8 A0A0N8B2C8 K1QEY5 A0A0P5TUW7 A0A0P5I8G9 A0A0P5DUB9 A0A0N8DKE9 A0A0P5R4A7 A0A0P5R7A7 A0A0P5Q991 A0A0P5YTS7 A0A131YG45 A0A0P5D2Z2 A0A0N8CDC2 A0A0P5DGS8 A0A224YXG4 A0A0P5VYU5 A0A0P5UKB3 A0A147BPD1 A0A0P5NJF4 A0A0P5G0C5 A0A0P5XCK9 A0A1Y1KUT7 A0A131XRY8 A0A139WH71 A0A0P5LCI5 A0A1W4X289 A0A0P6DP87 A0A0P5QEQ0 A0A1S4FUL6 Q16NN6 A0A182H367 A0A182PCA1 A0A182JPY1 A0A084WII2 A0A182J119 A0A182FB79 W5JW25 A0A0P5RS99 A0A182QLH9 A0A182Y9I2 A0A2M4A7V1 A0A182N2S7 A0A0U2UAP2 A0A336KUF3 A0A1L8DSE2 A0A182RVG4 A0A182LWW5 A0A182WE12 A0A1Q3FAY6 A0A146LRU8 A0A182HHM1 Q7PZI6 A0A182UTQ6 A0A023F2T6 A0A182L569 A0A0P6HXA4 A0A0P4VNN6 A0A182U438 A0A0A9WIY7 A0A0A1WVB5 A0A0V0G4N4 A0A069DZQ0 A0A1I8MXZ1
PDB
4XIB
E-value=0,
Score=1630
Ontologies
GO
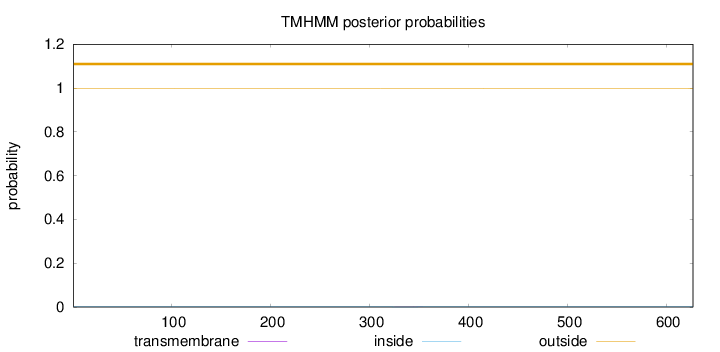
Topology
Length:
627
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00837999999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00065
outside
1 - 627
Population Genetic Test Statistics
Pi
266.172149
Theta
188.234526
Tajima's D
1.23188
CLR
0.006845
CSRT
0.722013899305035
Interpretation
Uncertain
