Gene
KWMTBOMO02068
Pre Gene Modal
BGIBMGA002976
Annotation
PREDICTED:_WW_domain-containing_oxidoreductase-like_isoform_X1_[Bombyx_mori]
Full name
WW domain-containing oxidoreductase
Location in the cell
Mitochondrial Reliability : 1.27
Sequence
CDS
ATGGCGACCGTCACAACTCGAAACCTCATTTTAAATGATAAAGTAAGAGCCTTTAGTTCTTCTAAAGTCAAATCTTTGGTTGAAAATTATTTTGATTTAAATTTTCGAATACACGGACCAACTGGCGAAGAGGTGACAAAAGGTCTAGATTTAAAAGATAAAACATGTTTGATAACTGGAGCGAACAGCGGCGTCGGTTTAGAAATGACGAAATGTTTAAATTTATGCGGATGCACTGTCCTCATGGCGTGCCGGAACACGTATGCTGCAAGCGTCGTCGCTAAAAATACATGCCTCAAACCCGATAGTCTACGACTTTACGAATTAAATCTTGCTTCACTTAGATCTGTGCGAAAATGCTCTGATGATCTTTTAAATAAAGAAAAAAAAATTGATATTGTTATCCTGAATGCTGGAATTTTCGGCTTACCTTGGACAGAGACTGTGGATAAATTGGAAATAACGTTTCAAGTCAACTATCTAAGCCAATATTATCTGCTCATGAATATCGAAGATATTCTTGCCGATAATGCAAGAGTCGTAATAATATCGTCAGAATCTCACAGATACGTAACGTTGCCTTTTAATGAAATATTAACGCCAACTGAGGAAACACTTTCACTTCCCAAAAATAAATATAGCTCCATCAAAGCATATAATATTTCAAAATTGTGTGGAATATTAGCCATGCATAATCTAGGATATCGTTGGCTAAACACTAAGAAAACTGTGATCAGCGCGCACCCGGGTTCATTTGTGAAAACAAGATTGTGTCGTAATTGGTGGCTTTACGAAGCTATCTACACCGGGATGAAACCGTTTACCAAAACTATAGCTCAAGCTGCTAGTACACCACTGTACTGCGCTACATCATCCGATTTGAATGGTTTATCGGCGATCTATTACAAGAATTGTGAACGTTGTCAAGAGAGCGACCTGGCTAGCAATCTCCAGTTATCGTTTCGAATAAACGATTTGACCAAAGACATTATTCGAGATAGAGTGACGATGTCAGAGTTGCCAGTTTCTTTGTCAGACACGCGTCAAAAACACGACGTCGAACACAATATCGAAGATTCACTTTTAACGAACTATTCAAATTGA
Protein
MATVTTRNLILNDKVRAFSSSKVKSLVENYFDLNFRIHGPTGEEVTKGLDLKDKTCLITGANSGVGLEMTKCLNLCGCTVLMACRNTYAASVVAKNTCLKPDSLRLYELNLASLRSVRKCSDDLLNKEKKIDIVILNAGIFGLPWTETVDKLEITFQVNYLSQYYLLMNIEDILADNARVVIISSESHRYVTLPFNEILTPTEETLSLPKNKYSSIKAYNISKLCGILAMHNLGYRWLNTKKTVISAHPGSFVKTRLCRNWWLYEAIYTGMKPFTKTIAQAASTPLYCATSSDLNGLSAIYYKNCERCQESDLASNLQLSFRINDLTKDIIRDRVTMSELPVSLSDTRQKHDVEHNIEDSLLTNYSN
Summary
Description
Putative oxidoreductase. May control genotoxic stress-induced cell death. May play a role in TGFB1 signaling and TGFB1-mediated cell death. May also play a role in tumor necrosis factor (TNF)-mediated cell death. May play a role in Wnt signaling (By similarity).
Putative oxidoreductase. Acts as a tumor suppressor and plays a role in apoptosis. May function synergistically with p53/TP53 to control genotoxic stress-induced cell death. Plays a role in TGFB1 signaling and TGFB1-mediated cell death. May also play a role in tumor necrosis factor (TNF)-mediated cell death. Required for normal bone development. Inhibits Wnt signaling (By similarity).
Putative oxidoreductase. Acts as a tumor suppressor and plays a role in apoptosis. May function synergistically with p53/TP53 to control genotoxic stress-induced cell death. Plays a role in TGFB1 signaling and TGFB1-mediated cell death. Inhibits Wnt signaling, probably by sequestering DVL2 in the cytoplasm (By similarity). May also play a role in tumor necrosis factor (TNF)-mediated cell death. Required for normal bone development.
Putative oxidoreductase. Acts as a tumor suppressor and plays a role in apoptosis. May function synergistically with p53/TP53 to control genotoxic stress-induced cell death. Plays a role in TGFB1 signaling and TGFB1-mediated cell death. May also play a role in tumor necrosis factor (TNF)-mediated cell death. Required for normal bone development. Inhibits Wnt signaling (By similarity).
Putative oxidoreductase. Acts as a tumor suppressor and plays a role in apoptosis. May function synergistically with p53/TP53 to control genotoxic stress-induced cell death. Plays a role in TGFB1 signaling and TGFB1-mediated cell death. Inhibits Wnt signaling, probably by sequestering DVL2 in the cytoplasm (By similarity). May also play a role in tumor necrosis factor (TNF)-mediated cell death. Required for normal bone development.
Subunit
Interacts with TP53, TP73/p73 and MAPK8. Interacts with MAPT/TAU. Forms a ternary complex with TP53 and MDM2 and interacts with ERBB4, LITAF and WBP1. May interact with FAM189B and SCOTIN (By similarity). Interacts with HYAL2 and RUNX2. Interacts with TNK2 (By similarity). Interacts with TMEM207 (By similarity).
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
Apoptosis
Complete proteome
Cytoplasm
NADP
Oxidoreductase
Phosphoprotein
Reference proteome
Repeat
Wnt signaling pathway
Nucleus
Alternative splicing
Mitochondrion
Tumor suppressor
Ubl conjugation
Feature
chain WW domain-containing oxidoreductase
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9J0E0
A0A2A4K6E4
A0A2H1WM68
A0A0L7LPV0
A0A212EKM0
A0A0M4EFS5
+ More
A0A1A9XKX5 A0A1B0BCB9 A0A1I8NS07 B4JAP5 A0A1B0FHX1 E2A102 A0A1B0A0P3 A0A151INK6 B4LTS9 A0A1J1IU20 A0A1I8MCZ5 A0A1W4W8U5 E2BKL5 A0A1A9WHK3 A0A151I5L6 X2J7F1 Q9VLU5 F4X276 B4KGA7 A0A0K8TKQ7 A0A026WCJ1 A0A195DRL7 A0A1L8E4S7 B3MJQ7 E9JCD3 B4Q681 B4HYC9 A0A182IC26 Q7Q4J0 A0A158NAK8 B3N6R6 Q29LV4 A0A151X589 A0A1B0DHY2 B4MVS4 B4G7D1 B4NWB6 A0A2M3Z8N1 A0A2M4ALH4 A0A3B0JAT8 A0A182RG27 A0A2M4CUW6 A0A2M4CUG5 A0A2M4BPW5 A0A1Y1LL89 A0A232EZ24 B0WCM6 A0A2A4K763 A0A0L0BZA8 A0A2J7PWP7 A0A1B6E0E8 A0A210PNE8 A0A194QPP0 A0A1W4X8N9 A0A212EYE6 K7IRI6 A0A0A9WL30 A0A2J7PWQ7 A0A194PHJ3 V4BFR0 A0A0A9WTG0 A0A182HAA8 A0A067R7F5 K1RNM4 Q1HQF3 Q17K82 A0A0L7RI52 A0A1D2N5P5 T1KXR1 A0A0B7AL24 A0A0F7Z311 A0A2C9JT57 A0A2T7NIQ0 A0A023EYB3 A0A1W5AUD6 A0A2H1X0A6 V9KUE4 A0A2R7VRT7 A0A1Q3FHM0 A0A1Q3FHK3 A0A182GME3 A0A2W1BS02 A0A0L8HL73 Q5F389 W4XH59 A0A088A8B9 A0A154PPU7 A0A069DTX2 A0A0V0G846 A0A087TND9 A0A087YI21 A0A3P9PLF4 A0A3B3U3D0 Q91WL8
A0A1A9XKX5 A0A1B0BCB9 A0A1I8NS07 B4JAP5 A0A1B0FHX1 E2A102 A0A1B0A0P3 A0A151INK6 B4LTS9 A0A1J1IU20 A0A1I8MCZ5 A0A1W4W8U5 E2BKL5 A0A1A9WHK3 A0A151I5L6 X2J7F1 Q9VLU5 F4X276 B4KGA7 A0A0K8TKQ7 A0A026WCJ1 A0A195DRL7 A0A1L8E4S7 B3MJQ7 E9JCD3 B4Q681 B4HYC9 A0A182IC26 Q7Q4J0 A0A158NAK8 B3N6R6 Q29LV4 A0A151X589 A0A1B0DHY2 B4MVS4 B4G7D1 B4NWB6 A0A2M3Z8N1 A0A2M4ALH4 A0A3B0JAT8 A0A182RG27 A0A2M4CUW6 A0A2M4CUG5 A0A2M4BPW5 A0A1Y1LL89 A0A232EZ24 B0WCM6 A0A2A4K763 A0A0L0BZA8 A0A2J7PWP7 A0A1B6E0E8 A0A210PNE8 A0A194QPP0 A0A1W4X8N9 A0A212EYE6 K7IRI6 A0A0A9WL30 A0A2J7PWQ7 A0A194PHJ3 V4BFR0 A0A0A9WTG0 A0A182HAA8 A0A067R7F5 K1RNM4 Q1HQF3 Q17K82 A0A0L7RI52 A0A1D2N5P5 T1KXR1 A0A0B7AL24 A0A0F7Z311 A0A2C9JT57 A0A2T7NIQ0 A0A023EYB3 A0A1W5AUD6 A0A2H1X0A6 V9KUE4 A0A2R7VRT7 A0A1Q3FHM0 A0A1Q3FHK3 A0A182GME3 A0A2W1BS02 A0A0L8HL73 Q5F389 W4XH59 A0A088A8B9 A0A154PPU7 A0A069DTX2 A0A0V0G846 A0A087TND9 A0A087YI21 A0A3P9PLF4 A0A3B3U3D0 Q91WL8
EC Number
1.1.1.-
Pubmed
19121390
26227816
22118469
17994087
20798317
25315136
+ More
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 16007179 21719571 26369729 24508170 30249741 21282665 22936249 12364791 14747013 17210077 21347285 15632085 17550304 28004739 28648823 26108605 28812685 26354079 20075255 25401762 26823975 23254933 26483478 24845553 22992520 17204158 17510324 27289101 15562597 25474469 24402279 28756777 15642098 26334808 11058590 16141072 15489334 11979549 12514174 15126504 15345747 15026124 16219768 15664696 17360458 18487609 19366691 21183079
10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 12537569 16007179 21719571 26369729 24508170 30249741 21282665 22936249 12364791 14747013 17210077 21347285 15632085 17550304 28004739 28648823 26108605 28812685 26354079 20075255 25401762 26823975 23254933 26483478 24845553 22992520 17204158 17510324 27289101 15562597 25474469 24402279 28756777 15642098 26334808 11058590 16141072 15489334 11979549 12514174 15126504 15345747 15026124 16219768 15664696 17360458 18487609 19366691 21183079
EMBL
BABH01010530
NWSH01000081
PCG79811.1
ODYU01009616
SOQ54173.1
JTDY01000385
+ More
KOB77434.1 AGBW02014274 OWR41998.1 CP012523 ALC39367.1 JXJN01011917 JXJN01011918 CH916368 EDW03853.1 CCAG010002083 CCAG010002084 GL435626 EFN73055.1 KQ976932 KYN07046.1 CH940649 EDW63980.1 CVRI01000059 CRL03608.1 GL448819 EFN83713.1 KQ976413 KYM90378.1 AE014134 AHN54249.1 AY119574 GL888565 EGI59492.1 CH933807 EDW11094.1 GDAI01002699 JAI14904.1 KK107276 QOIP01000006 EZA53638.1 RLU21110.1 KQ980612 KYN15144.1 GFDF01000418 JAV13666.1 CH902620 EDV31396.1 GL771786 EFZ09538.1 CM000361 CM002910 EDX04157.1 KMY88893.1 CH480818 EDW52059.1 APCN01000774 AAAB01008964 EAA12850.4 ADTU01010368 CH954177 EDV59282.1 CH379060 EAL33941.2 KQ982519 KYQ55566.1 AJVK01006048 AJVK01006049 CH963857 EDW75794.2 CH479180 EDW29264.1 CM000157 EDW88433.1 GGFM01004136 MBW24887.1 GGFK01008310 MBW41631.1 OUUW01000004 SPP79444.1 GGFL01004783 MBW68961.1 GGFL01004782 MBW68960.1 GGFJ01005862 MBW55003.1 GEZM01052549 JAV74444.1 NNAY01001561 OXU23593.1 DS231889 EDS43677.1 NWSH01000098 PCG79603.1 JRES01001119 KNC25397.1 NEVH01020869 PNF20763.1 GEDC01008769 GEDC01005903 JAS28529.1 JAS31395.1 NEDP02005575 OWF38011.1 KQ461194 KPJ06945.1 AGBW02011531 OWR46512.1 GBHO01035508 GBHO01035507 GBHO01035506 GBRD01002304 GBRD01002302 GDHC01020892 JAG08096.1 JAG08097.1 JAG08098.1 JAG63517.1 JAP97736.1 PNF20761.1 KQ459603 KPI92886.1 KB202823 ESO87819.1 GBHO01035504 GBHO01035503 GBRD01002303 GDHC01002727 GDHC01000239 JAG08100.1 JAG08101.1 JAG63518.1 JAQ15902.1 JAQ18390.1 JXUM01032067 KQ560944 KXJ80241.1 KK852655 KDR19283.1 JH815822 EKC35996.1 DQ440491 CH477468 ABF18524.1 EAT40479.1 CH477227 EAT47103.1 KQ414584 KOC70667.1 LJIJ01000210 ODN00385.1 CAEY01000696 HACG01034879 CEK81744.1 GBEX01004012 JAI10548.1 PZQS01000012 PVD21049.1 GBBI01004397 JAC14315.1 ODYU01012422 SOQ58717.1 JW869638 AFP02156.1 KK854015 PTY09390.1 GFDL01008023 JAV27022.1 GFDL01008017 JAV27028.1 JXUM01014080 KQ560393 KXJ82655.1 KZ149975 PZC75947.1 KQ417888 KOF89967.1 AJ851761 AAGJ04048706 AAGJ04048707 AAGJ04048708 KQ435022 KZC13916.1 GBGD01001668 JAC87221.1 GECL01001868 JAP04256.1 KK116036 KFM66628.1 AYCK01001909 AYCK01001910 AYCK01001911 AYCK01001912 AF187014 AK018507 AK019911 AK046903 AK078528 BC014716 AH011063
KOB77434.1 AGBW02014274 OWR41998.1 CP012523 ALC39367.1 JXJN01011917 JXJN01011918 CH916368 EDW03853.1 CCAG010002083 CCAG010002084 GL435626 EFN73055.1 KQ976932 KYN07046.1 CH940649 EDW63980.1 CVRI01000059 CRL03608.1 GL448819 EFN83713.1 KQ976413 KYM90378.1 AE014134 AHN54249.1 AY119574 GL888565 EGI59492.1 CH933807 EDW11094.1 GDAI01002699 JAI14904.1 KK107276 QOIP01000006 EZA53638.1 RLU21110.1 KQ980612 KYN15144.1 GFDF01000418 JAV13666.1 CH902620 EDV31396.1 GL771786 EFZ09538.1 CM000361 CM002910 EDX04157.1 KMY88893.1 CH480818 EDW52059.1 APCN01000774 AAAB01008964 EAA12850.4 ADTU01010368 CH954177 EDV59282.1 CH379060 EAL33941.2 KQ982519 KYQ55566.1 AJVK01006048 AJVK01006049 CH963857 EDW75794.2 CH479180 EDW29264.1 CM000157 EDW88433.1 GGFM01004136 MBW24887.1 GGFK01008310 MBW41631.1 OUUW01000004 SPP79444.1 GGFL01004783 MBW68961.1 GGFL01004782 MBW68960.1 GGFJ01005862 MBW55003.1 GEZM01052549 JAV74444.1 NNAY01001561 OXU23593.1 DS231889 EDS43677.1 NWSH01000098 PCG79603.1 JRES01001119 KNC25397.1 NEVH01020869 PNF20763.1 GEDC01008769 GEDC01005903 JAS28529.1 JAS31395.1 NEDP02005575 OWF38011.1 KQ461194 KPJ06945.1 AGBW02011531 OWR46512.1 GBHO01035508 GBHO01035507 GBHO01035506 GBRD01002304 GBRD01002302 GDHC01020892 JAG08096.1 JAG08097.1 JAG08098.1 JAG63517.1 JAP97736.1 PNF20761.1 KQ459603 KPI92886.1 KB202823 ESO87819.1 GBHO01035504 GBHO01035503 GBRD01002303 GDHC01002727 GDHC01000239 JAG08100.1 JAG08101.1 JAG63518.1 JAQ15902.1 JAQ18390.1 JXUM01032067 KQ560944 KXJ80241.1 KK852655 KDR19283.1 JH815822 EKC35996.1 DQ440491 CH477468 ABF18524.1 EAT40479.1 CH477227 EAT47103.1 KQ414584 KOC70667.1 LJIJ01000210 ODN00385.1 CAEY01000696 HACG01034879 CEK81744.1 GBEX01004012 JAI10548.1 PZQS01000012 PVD21049.1 GBBI01004397 JAC14315.1 ODYU01012422 SOQ58717.1 JW869638 AFP02156.1 KK854015 PTY09390.1 GFDL01008023 JAV27022.1 GFDL01008017 JAV27028.1 JXUM01014080 KQ560393 KXJ82655.1 KZ149975 PZC75947.1 KQ417888 KOF89967.1 AJ851761 AAGJ04048706 AAGJ04048707 AAGJ04048708 KQ435022 KZC13916.1 GBGD01001668 JAC87221.1 GECL01001868 JAP04256.1 KK116036 KFM66628.1 AYCK01001909 AYCK01001910 AYCK01001911 AYCK01001912 AF187014 AK018507 AK019911 AK046903 AK078528 BC014716 AH011063
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000092553
UP000092443
+ More
UP000092460 UP000095300 UP000001070 UP000092444 UP000000311 UP000092445 UP000078542 UP000008792 UP000183832 UP000095301 UP000192221 UP000008237 UP000091820 UP000078540 UP000000803 UP000007755 UP000009192 UP000053097 UP000279307 UP000078492 UP000007801 UP000000304 UP000001292 UP000075840 UP000007062 UP000005205 UP000008711 UP000001819 UP000075809 UP000092462 UP000007798 UP000008744 UP000002282 UP000268350 UP000075900 UP000215335 UP000002320 UP000037069 UP000235965 UP000242188 UP000053240 UP000192223 UP000002358 UP000053268 UP000030746 UP000069940 UP000249989 UP000027135 UP000005408 UP000008820 UP000053825 UP000094527 UP000015104 UP000076420 UP000245119 UP000192224 UP000053454 UP000000539 UP000007110 UP000005203 UP000076502 UP000054359 UP000028760 UP000242638 UP000261500 UP000000589
UP000092460 UP000095300 UP000001070 UP000092444 UP000000311 UP000092445 UP000078542 UP000008792 UP000183832 UP000095301 UP000192221 UP000008237 UP000091820 UP000078540 UP000000803 UP000007755 UP000009192 UP000053097 UP000279307 UP000078492 UP000007801 UP000000304 UP000001292 UP000075840 UP000007062 UP000005205 UP000008711 UP000001819 UP000075809 UP000092462 UP000007798 UP000008744 UP000002282 UP000268350 UP000075900 UP000215335 UP000002320 UP000037069 UP000235965 UP000242188 UP000053240 UP000192223 UP000002358 UP000053268 UP000030746 UP000069940 UP000249989 UP000027135 UP000005408 UP000008820 UP000053825 UP000094527 UP000015104 UP000076420 UP000245119 UP000192224 UP000053454 UP000000539 UP000007110 UP000005203 UP000076502 UP000054359 UP000028760 UP000242638 UP000261500 UP000000589
Interpro
CDD
ProteinModelPortal
H9J0E0
A0A2A4K6E4
A0A2H1WM68
A0A0L7LPV0
A0A212EKM0
A0A0M4EFS5
+ More
A0A1A9XKX5 A0A1B0BCB9 A0A1I8NS07 B4JAP5 A0A1B0FHX1 E2A102 A0A1B0A0P3 A0A151INK6 B4LTS9 A0A1J1IU20 A0A1I8MCZ5 A0A1W4W8U5 E2BKL5 A0A1A9WHK3 A0A151I5L6 X2J7F1 Q9VLU5 F4X276 B4KGA7 A0A0K8TKQ7 A0A026WCJ1 A0A195DRL7 A0A1L8E4S7 B3MJQ7 E9JCD3 B4Q681 B4HYC9 A0A182IC26 Q7Q4J0 A0A158NAK8 B3N6R6 Q29LV4 A0A151X589 A0A1B0DHY2 B4MVS4 B4G7D1 B4NWB6 A0A2M3Z8N1 A0A2M4ALH4 A0A3B0JAT8 A0A182RG27 A0A2M4CUW6 A0A2M4CUG5 A0A2M4BPW5 A0A1Y1LL89 A0A232EZ24 B0WCM6 A0A2A4K763 A0A0L0BZA8 A0A2J7PWP7 A0A1B6E0E8 A0A210PNE8 A0A194QPP0 A0A1W4X8N9 A0A212EYE6 K7IRI6 A0A0A9WL30 A0A2J7PWQ7 A0A194PHJ3 V4BFR0 A0A0A9WTG0 A0A182HAA8 A0A067R7F5 K1RNM4 Q1HQF3 Q17K82 A0A0L7RI52 A0A1D2N5P5 T1KXR1 A0A0B7AL24 A0A0F7Z311 A0A2C9JT57 A0A2T7NIQ0 A0A023EYB3 A0A1W5AUD6 A0A2H1X0A6 V9KUE4 A0A2R7VRT7 A0A1Q3FHM0 A0A1Q3FHK3 A0A182GME3 A0A2W1BS02 A0A0L8HL73 Q5F389 W4XH59 A0A088A8B9 A0A154PPU7 A0A069DTX2 A0A0V0G846 A0A087TND9 A0A087YI21 A0A3P9PLF4 A0A3B3U3D0 Q91WL8
A0A1A9XKX5 A0A1B0BCB9 A0A1I8NS07 B4JAP5 A0A1B0FHX1 E2A102 A0A1B0A0P3 A0A151INK6 B4LTS9 A0A1J1IU20 A0A1I8MCZ5 A0A1W4W8U5 E2BKL5 A0A1A9WHK3 A0A151I5L6 X2J7F1 Q9VLU5 F4X276 B4KGA7 A0A0K8TKQ7 A0A026WCJ1 A0A195DRL7 A0A1L8E4S7 B3MJQ7 E9JCD3 B4Q681 B4HYC9 A0A182IC26 Q7Q4J0 A0A158NAK8 B3N6R6 Q29LV4 A0A151X589 A0A1B0DHY2 B4MVS4 B4G7D1 B4NWB6 A0A2M3Z8N1 A0A2M4ALH4 A0A3B0JAT8 A0A182RG27 A0A2M4CUW6 A0A2M4CUG5 A0A2M4BPW5 A0A1Y1LL89 A0A232EZ24 B0WCM6 A0A2A4K763 A0A0L0BZA8 A0A2J7PWP7 A0A1B6E0E8 A0A210PNE8 A0A194QPP0 A0A1W4X8N9 A0A212EYE6 K7IRI6 A0A0A9WL30 A0A2J7PWQ7 A0A194PHJ3 V4BFR0 A0A0A9WTG0 A0A182HAA8 A0A067R7F5 K1RNM4 Q1HQF3 Q17K82 A0A0L7RI52 A0A1D2N5P5 T1KXR1 A0A0B7AL24 A0A0F7Z311 A0A2C9JT57 A0A2T7NIQ0 A0A023EYB3 A0A1W5AUD6 A0A2H1X0A6 V9KUE4 A0A2R7VRT7 A0A1Q3FHM0 A0A1Q3FHK3 A0A182GME3 A0A2W1BS02 A0A0L8HL73 Q5F389 W4XH59 A0A088A8B9 A0A154PPU7 A0A069DTX2 A0A0V0G846 A0A087TND9 A0A087YI21 A0A3P9PLF4 A0A3B3U3D0 Q91WL8
PDB
3RD5
E-value=9.69867e-15,
Score=195
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus
Mitochondrion Partially localizes to the mitochondria (By similarity). Translocates to the nucleus in response to TGFB1 (PubMed:19366691). Translocates to the nucleus upon genotoxic stress or TNF stimulation (PubMed:11058590). With evidence from 3 publications.
Nucleus
Mitochondrion Partially localizes to the mitochondria (By similarity). Translocates to the nucleus in response to TGFB1 (PubMed:19366691). Translocates to the nucleus upon genotoxic stress or TNF stimulation (PubMed:11058590). With evidence from 3 publications.
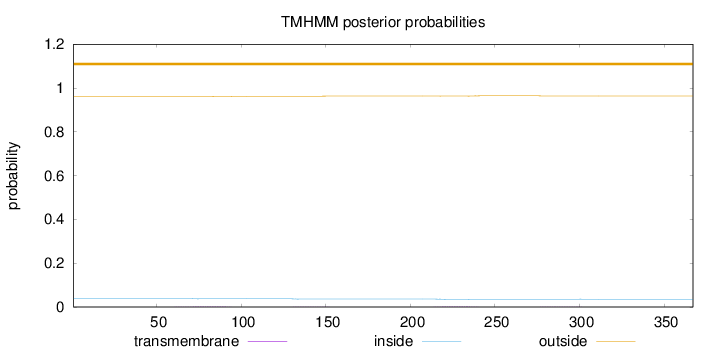
Length:
367
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10573
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.03901
outside
1 - 367
Population Genetic Test Statistics
Pi
215.11626
Theta
180.592557
Tajima's D
1.083757
CLR
0.021385
CSRT
0.687415629218539
Interpretation
Uncertain
