Gene
KWMTBOMO02066 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002975
Annotation
putative_39S_ribosomal_protein_L15?_mitochondrial_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.246 Mitochondrial Reliability : 1.867
Sequence
CDS
ATGGGTTCGCGGCAAATAACAGAGAAATCACTGTCTATGTTGAGGTCGTTACCAAGAATCACGCTAGCAAATATTCGTGATAATCCGGGGTCTAAAATAACACAAAAAAGAGGAAGGGGTCAACACGGGGGTGACAAACATGGTGCAGGCAATAAAGGTTCTGGACAACGTCAAAATTATATGAGACTAGGTTATGAAACTGGAAACAACCCATTTTATCTTCGTATTCCTCATGAGAATTACTATAGAGGACACCATTTGAAAAGAGAGTATCCTCCTATGACACTTTTGGAGTTACAAAAATTGATCGATAGAAACCGAGTAGACATATCTAAACCCATTGATATTGCCAGCTTGATAAAATCTGGGTTATACACATTGAATCCTGACCAAAAGCATTTTGGTGTTAATTTAACTGATGATGGAGCTGACATATTTGCTGCAAAAATTAATATAGAAGTACAGTGGGCAAGTGAACAAGTTATTGCAGCGATTGAGAAAAATGGTGGTGTTATTACTACAGCCTACTATGATCCTCACAGTTTATTTATATTGAAAAATCCTCAAAGGTTTTTTGAAACTGGTCAAGCTATTCCAAGAAGAATGATTCCTCCGCCCGATGCCATTGAATTTTACACCAGTGCTGCAACACGTGGTTATTTAGCAGATCCAGAAAAAATATCTCAAGAAAGATTAAAATTGTCTCAGAAATATGGATATACACTTCCAAATTTAGAAAGTGACCCACAATATGAGATGTTAACAGATAGAAAGGATCCTAGACAAATATTTTTTGGATTACAGCCAGGGTGGGTAATTAATTTGAAAGATGAATGCATTTTAAAGCCAAAAGATGAAGAATTATTACAGTTCTATTCTATGTAG
Protein
MGSRQITEKSLSMLRSLPRITLANIRDNPGSKITQKRGRGQHGGDKHGAGNKGSGQRQNYMRLGYETGNNPFYLRIPHENYYRGHHLKREYPPMTLLELQKLIDRNRVDISKPIDIASLIKSGLYTLNPDQKHFGVNLTDDGADIFAAKINIEVQWASEQVIAAIEKNGGVITTAYYDPHSLFILKNPQRFFETGQAIPRRMIPPPDAIEFYTSAATRGYLADPEKISQERLKLSQKYGYTLPNLESDPQYEMLTDRKDPRQIFFGLQPGWVINLKDECILKPKDEELLQFYSM
Summary
Uniprot
H9J0D9
A0A1E1WSF6
A0A194QNF0
A0A212EKA7
A0A2A4K6U8
A0A2H1VD17
+ More
A0A194PJY0 A0A0L7LPS9 A0A2P8ZMQ9 A0A067QIX9 A0A1Y1M6E8 A0A1B6HB62 A0A1B6K8J4 A0A1B6FVE4 A0A151XE55 A0A195BIN3 A0A310SPH6 E9JCD2 D6WM14 F4WIJ6 A0A195F533 A0A195DZ42 A0A158NJZ4 A0A1Q3FB77 B0WLQ4 A0A088A2E6 E2A101 A0A182T6C2 A0A182PW34 A0A154PLV1 A0A2A3EBE4 A0A182IV89 A0A1I8MA29 A0A182QY69 A0A182N2R3 T1PIL0 A0A182RXU8 A0A2S2RAY1 A0A2M4ASC8 V5GVZ4 A0A1E1WR07 A0A2M4BVY3 A0A1I8NW16 A0A182Y9J5 A0A240SY96 A0A1B6CJR2 A0A182WE24 A0A2M3Z9X0 A0A232EYN0 A0A182USL5 A0A182UBJ4 A0A182XJQ1 Q7PZJ6 A0A182HHN1 A0A0L7R3C9 A0A084WIG0 K7IRI5 A0A182MTA7 A0A2M3ZIK6 A0A1S4F4W9 W5JWQ4 A0A182JY21 A0A182FB89 A0A182L582 A0A182HEI8 Q17FD8 A0A1B0CEF4 B4KY38 A0A026WC49 A0A1A9WH57 Q8T9L3 A0A1A9YCL0 A0A1B0AQM5 B4LGD2 B3NIM1 A0A2H8TTY4 C4WUA1 A0A0Q9WTI2 A0A0M4EC12 U5ERU0 N6UF61 Q9VPF6 B4IUG5 A0A1L8DRJ1 B4IAA9 A0A1W4VV74 A0A034VKE3 B4QRZ5 W8BYK6 A0A1L8DRT5 A0A034VL82 B3M998 A0A1A9V558 A0A0L0BL13 J9JST4 D3TMK5 B4N6M8 A0A1A9ZWG0 A0A0P5KFS6 A0A2S2NIT4
A0A194PJY0 A0A0L7LPS9 A0A2P8ZMQ9 A0A067QIX9 A0A1Y1M6E8 A0A1B6HB62 A0A1B6K8J4 A0A1B6FVE4 A0A151XE55 A0A195BIN3 A0A310SPH6 E9JCD2 D6WM14 F4WIJ6 A0A195F533 A0A195DZ42 A0A158NJZ4 A0A1Q3FB77 B0WLQ4 A0A088A2E6 E2A101 A0A182T6C2 A0A182PW34 A0A154PLV1 A0A2A3EBE4 A0A182IV89 A0A1I8MA29 A0A182QY69 A0A182N2R3 T1PIL0 A0A182RXU8 A0A2S2RAY1 A0A2M4ASC8 V5GVZ4 A0A1E1WR07 A0A2M4BVY3 A0A1I8NW16 A0A182Y9J5 A0A240SY96 A0A1B6CJR2 A0A182WE24 A0A2M3Z9X0 A0A232EYN0 A0A182USL5 A0A182UBJ4 A0A182XJQ1 Q7PZJ6 A0A182HHN1 A0A0L7R3C9 A0A084WIG0 K7IRI5 A0A182MTA7 A0A2M3ZIK6 A0A1S4F4W9 W5JWQ4 A0A182JY21 A0A182FB89 A0A182L582 A0A182HEI8 Q17FD8 A0A1B0CEF4 B4KY38 A0A026WC49 A0A1A9WH57 Q8T9L3 A0A1A9YCL0 A0A1B0AQM5 B4LGD2 B3NIM1 A0A2H8TTY4 C4WUA1 A0A0Q9WTI2 A0A0M4EC12 U5ERU0 N6UF61 Q9VPF6 B4IUG5 A0A1L8DRJ1 B4IAA9 A0A1W4VV74 A0A034VKE3 B4QRZ5 W8BYK6 A0A1L8DRT5 A0A034VL82 B3M998 A0A1A9V558 A0A0L0BL13 J9JST4 D3TMK5 B4N6M8 A0A1A9ZWG0 A0A0P5KFS6 A0A2S2NIT4
Pubmed
19121390
26354079
22118469
26227816
29403074
24845553
+ More
28004739 21282665 18362917 19820115 21719571 21347285 20798317 25315136 25244985 28648823 12364791 14747013 17210077 24438588 20075255 20920257 23761445 20966253 26483478 17510324 17994087 24508170 30249741 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 22936249 24495485 26108605 20353571
28004739 21282665 18362917 19820115 21719571 21347285 20798317 25315136 25244985 28648823 12364791 14747013 17210077 24438588 20075255 20920257 23761445 20966253 26483478 17510324 17994087 24508170 30249741 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 22936249 24495485 26108605 20353571
EMBL
BABH01010532
GDQN01001075
JAT89979.1
KQ461196
KPJ06500.1
AGBW02014319
+ More
OWR41878.1 NWSH01000081 PCG79799.1 ODYU01001874 SOQ38686.1 KQ459601 KPI93741.1 JTDY01000385 KOB77442.1 PYGN01000015 PSN57786.1 KK853470 KDR07389.1 GEZM01039146 JAV81409.1 GECU01035874 JAS71832.1 GEBQ01032209 JAT07768.1 GECZ01015635 JAS54134.1 KQ982254 KYQ58652.1 KQ976465 KYM84271.1 KQ761976 OAD56420.1 GL771786 EFZ09535.1 KQ971343 EFA04204.1 GL888176 EGI65932.1 KQ981820 KYN35282.1 KQ980031 KYN18160.1 ADTU01018485 GFDL01010235 JAV24810.1 DS231989 EDS30619.1 GL435626 EFN73054.1 KQ434972 KZC12737.1 KZ288311 PBC28506.1 AXCN02001086 KA647768 AFP62397.1 GGMS01017915 MBY87118.1 GGFK01010363 MBW43684.1 GALX01000137 JAB68329.1 GDQN01001646 JAT89408.1 GGFJ01008094 MBW57235.1 AJVK01024477 GEDC01023705 GEDC01015880 JAS13593.1 JAS21418.1 GGFM01004573 MBW25324.1 NNAY01001561 OXU23596.1 AAAB01008986 EAA00259.1 APCN01002431 KQ414663 KOC65377.1 ATLV01023931 KE525347 KFB50004.1 AXCM01003350 GGFM01007581 MBW28332.1 ADMH02000120 ETN67700.1 JXUM01131694 KQ567745 KXJ69300.1 CH477271 EAT45307.1 AJWK01008837 CH933809 EDW18740.1 KK107276 QOIP01000006 EZA53635.1 RLU21393.1 AY069243 AAL39388.1 JXJN01001971 CH940647 EDW70461.1 CH954178 EDV52517.1 GFXV01005909 MBW17714.1 AK340979 BAH71471.1 KRF84929.1 CP012525 ALC45104.1 GANO01002679 JAB57192.1 APGK01037326 KB740948 KB632288 ENN77297.1 ERL91556.1 AE014296 BT050572 AAF51599.1 ACJ23456.1 CH891902 EDX00029.1 GFDF01004998 JAV09086.1 CH480826 EDW44222.1 GAKP01016365 JAC42587.1 CM000363 CM002912 EDX11241.1 KMZ00791.1 GAMC01012214 GAMC01012212 GAMC01012210 GAMC01012208 GAMC01012206 GAMC01012204 JAB94349.1 GFDF01004997 JAV09087.1 GAKP01016372 GAKP01016368 GAKP01016363 JAC42580.1 CH902618 EDV40082.1 JRES01001711 KNC20633.1 ABLF02031777 ABLF02031790 CCAG010007514 EZ422657 ADD18933.1 CH964161 EDW80017.1 GDIQ01191481 JAK60244.1 GGMR01004419 MBY17038.1
OWR41878.1 NWSH01000081 PCG79799.1 ODYU01001874 SOQ38686.1 KQ459601 KPI93741.1 JTDY01000385 KOB77442.1 PYGN01000015 PSN57786.1 KK853470 KDR07389.1 GEZM01039146 JAV81409.1 GECU01035874 JAS71832.1 GEBQ01032209 JAT07768.1 GECZ01015635 JAS54134.1 KQ982254 KYQ58652.1 KQ976465 KYM84271.1 KQ761976 OAD56420.1 GL771786 EFZ09535.1 KQ971343 EFA04204.1 GL888176 EGI65932.1 KQ981820 KYN35282.1 KQ980031 KYN18160.1 ADTU01018485 GFDL01010235 JAV24810.1 DS231989 EDS30619.1 GL435626 EFN73054.1 KQ434972 KZC12737.1 KZ288311 PBC28506.1 AXCN02001086 KA647768 AFP62397.1 GGMS01017915 MBY87118.1 GGFK01010363 MBW43684.1 GALX01000137 JAB68329.1 GDQN01001646 JAT89408.1 GGFJ01008094 MBW57235.1 AJVK01024477 GEDC01023705 GEDC01015880 JAS13593.1 JAS21418.1 GGFM01004573 MBW25324.1 NNAY01001561 OXU23596.1 AAAB01008986 EAA00259.1 APCN01002431 KQ414663 KOC65377.1 ATLV01023931 KE525347 KFB50004.1 AXCM01003350 GGFM01007581 MBW28332.1 ADMH02000120 ETN67700.1 JXUM01131694 KQ567745 KXJ69300.1 CH477271 EAT45307.1 AJWK01008837 CH933809 EDW18740.1 KK107276 QOIP01000006 EZA53635.1 RLU21393.1 AY069243 AAL39388.1 JXJN01001971 CH940647 EDW70461.1 CH954178 EDV52517.1 GFXV01005909 MBW17714.1 AK340979 BAH71471.1 KRF84929.1 CP012525 ALC45104.1 GANO01002679 JAB57192.1 APGK01037326 KB740948 KB632288 ENN77297.1 ERL91556.1 AE014296 BT050572 AAF51599.1 ACJ23456.1 CH891902 EDX00029.1 GFDF01004998 JAV09086.1 CH480826 EDW44222.1 GAKP01016365 JAC42587.1 CM000363 CM002912 EDX11241.1 KMZ00791.1 GAMC01012214 GAMC01012212 GAMC01012210 GAMC01012208 GAMC01012206 GAMC01012204 JAB94349.1 GFDF01004997 JAV09087.1 GAKP01016372 GAKP01016368 GAKP01016363 JAC42580.1 CH902618 EDV40082.1 JRES01001711 KNC20633.1 ABLF02031777 ABLF02031790 CCAG010007514 EZ422657 ADD18933.1 CH964161 EDW80017.1 GDIQ01191481 JAK60244.1 GGMR01004419 MBY17038.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000053268
UP000037510
+ More
UP000245037 UP000027135 UP000075809 UP000078540 UP000007266 UP000007755 UP000078541 UP000078492 UP000005205 UP000002320 UP000005203 UP000000311 UP000075901 UP000075885 UP000076502 UP000242457 UP000075880 UP000095301 UP000075886 UP000075884 UP000075900 UP000095300 UP000076408 UP000092462 UP000075920 UP000215335 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000053825 UP000030765 UP000002358 UP000075883 UP000000673 UP000075881 UP000069272 UP000075882 UP000069940 UP000249989 UP000008820 UP000092461 UP000009192 UP000053097 UP000279307 UP000091820 UP000092443 UP000092460 UP000008792 UP000008711 UP000092553 UP000019118 UP000030742 UP000000803 UP000002282 UP000001292 UP000192221 UP000000304 UP000007801 UP000078200 UP000037069 UP000007819 UP000092444 UP000007798 UP000092445
UP000245037 UP000027135 UP000075809 UP000078540 UP000007266 UP000007755 UP000078541 UP000078492 UP000005205 UP000002320 UP000005203 UP000000311 UP000075901 UP000075885 UP000076502 UP000242457 UP000075880 UP000095301 UP000075886 UP000075884 UP000075900 UP000095300 UP000076408 UP000092462 UP000075920 UP000215335 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000053825 UP000030765 UP000002358 UP000075883 UP000000673 UP000075881 UP000069272 UP000075882 UP000069940 UP000249989 UP000008820 UP000092461 UP000009192 UP000053097 UP000279307 UP000091820 UP000092443 UP000092460 UP000008792 UP000008711 UP000092553 UP000019118 UP000030742 UP000000803 UP000002282 UP000001292 UP000192221 UP000000304 UP000007801 UP000078200 UP000037069 UP000007819 UP000092444 UP000007798 UP000092445
Pfam
PF00828 Ribosomal_L27A
Interpro
SUPFAM
SSF52080
SSF52080
ProteinModelPortal
H9J0D9
A0A1E1WSF6
A0A194QNF0
A0A212EKA7
A0A2A4K6U8
A0A2H1VD17
+ More
A0A194PJY0 A0A0L7LPS9 A0A2P8ZMQ9 A0A067QIX9 A0A1Y1M6E8 A0A1B6HB62 A0A1B6K8J4 A0A1B6FVE4 A0A151XE55 A0A195BIN3 A0A310SPH6 E9JCD2 D6WM14 F4WIJ6 A0A195F533 A0A195DZ42 A0A158NJZ4 A0A1Q3FB77 B0WLQ4 A0A088A2E6 E2A101 A0A182T6C2 A0A182PW34 A0A154PLV1 A0A2A3EBE4 A0A182IV89 A0A1I8MA29 A0A182QY69 A0A182N2R3 T1PIL0 A0A182RXU8 A0A2S2RAY1 A0A2M4ASC8 V5GVZ4 A0A1E1WR07 A0A2M4BVY3 A0A1I8NW16 A0A182Y9J5 A0A240SY96 A0A1B6CJR2 A0A182WE24 A0A2M3Z9X0 A0A232EYN0 A0A182USL5 A0A182UBJ4 A0A182XJQ1 Q7PZJ6 A0A182HHN1 A0A0L7R3C9 A0A084WIG0 K7IRI5 A0A182MTA7 A0A2M3ZIK6 A0A1S4F4W9 W5JWQ4 A0A182JY21 A0A182FB89 A0A182L582 A0A182HEI8 Q17FD8 A0A1B0CEF4 B4KY38 A0A026WC49 A0A1A9WH57 Q8T9L3 A0A1A9YCL0 A0A1B0AQM5 B4LGD2 B3NIM1 A0A2H8TTY4 C4WUA1 A0A0Q9WTI2 A0A0M4EC12 U5ERU0 N6UF61 Q9VPF6 B4IUG5 A0A1L8DRJ1 B4IAA9 A0A1W4VV74 A0A034VKE3 B4QRZ5 W8BYK6 A0A1L8DRT5 A0A034VL82 B3M998 A0A1A9V558 A0A0L0BL13 J9JST4 D3TMK5 B4N6M8 A0A1A9ZWG0 A0A0P5KFS6 A0A2S2NIT4
A0A194PJY0 A0A0L7LPS9 A0A2P8ZMQ9 A0A067QIX9 A0A1Y1M6E8 A0A1B6HB62 A0A1B6K8J4 A0A1B6FVE4 A0A151XE55 A0A195BIN3 A0A310SPH6 E9JCD2 D6WM14 F4WIJ6 A0A195F533 A0A195DZ42 A0A158NJZ4 A0A1Q3FB77 B0WLQ4 A0A088A2E6 E2A101 A0A182T6C2 A0A182PW34 A0A154PLV1 A0A2A3EBE4 A0A182IV89 A0A1I8MA29 A0A182QY69 A0A182N2R3 T1PIL0 A0A182RXU8 A0A2S2RAY1 A0A2M4ASC8 V5GVZ4 A0A1E1WR07 A0A2M4BVY3 A0A1I8NW16 A0A182Y9J5 A0A240SY96 A0A1B6CJR2 A0A182WE24 A0A2M3Z9X0 A0A232EYN0 A0A182USL5 A0A182UBJ4 A0A182XJQ1 Q7PZJ6 A0A182HHN1 A0A0L7R3C9 A0A084WIG0 K7IRI5 A0A182MTA7 A0A2M3ZIK6 A0A1S4F4W9 W5JWQ4 A0A182JY21 A0A182FB89 A0A182L582 A0A182HEI8 Q17FD8 A0A1B0CEF4 B4KY38 A0A026WC49 A0A1A9WH57 Q8T9L3 A0A1A9YCL0 A0A1B0AQM5 B4LGD2 B3NIM1 A0A2H8TTY4 C4WUA1 A0A0Q9WTI2 A0A0M4EC12 U5ERU0 N6UF61 Q9VPF6 B4IUG5 A0A1L8DRJ1 B4IAA9 A0A1W4VV74 A0A034VKE3 B4QRZ5 W8BYK6 A0A1L8DRT5 A0A034VL82 B3M998 A0A1A9V558 A0A0L0BL13 J9JST4 D3TMK5 B4N6M8 A0A1A9ZWG0 A0A0P5KFS6 A0A2S2NIT4
PDB
6NU3
E-value=1.19635e-79,
Score=753
Ontologies
GO
PANTHER
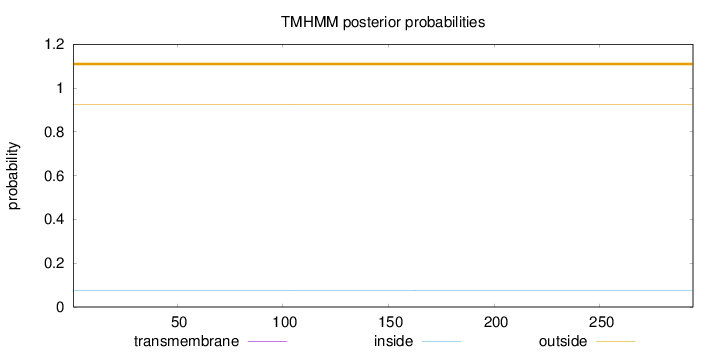
Topology
Length:
294
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000750000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07503
outside
1 - 294
Population Genetic Test Statistics
Pi
160.297832
Theta
147.992421
Tajima's D
0.69282
CLR
0.077131
CSRT
0.576171191440428
Interpretation
Uncertain
