Gene
KWMTBOMO02064 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003196
Annotation
VA0D_MANSE_RecName:_Full%3DV-type_proton_ATPase_subunit_d;_short=V-ATPase_subunit_d;_altname:_full=M40,V-ATPase_40_kDa_subunit,Vacuolar_proton_pump_subunit_d
Full name
V-type proton ATPase subunit
+ More
V-type proton ATPase subunit d
V-type proton ATPase subunit d 1
V-type proton ATPase subunit d
V-type proton ATPase subunit d 1
Alternative Name
M40
V-ATPase 40 kDa subunit
Vacuolar proton pump subunit d
V-ATPase 39 kDa subunit
Vacuolar H+ ATPase subunit AC39-1
Vacuolar proton pump subunit d 1
V-ATPase 40 kDa subunit
Vacuolar proton pump subunit d
V-ATPase 39 kDa subunit
Vacuolar H+ ATPase subunit AC39-1
Vacuolar proton pump subunit d 1
Location in the cell
Cytoplasmic Reliability : 2.696
Sequence
CDS
ATGAAGGGTTGCATATTTAATATTGATGCTGGGTATTTGGAGGGCTTGTGCCGTGGTTTCAAATGTGGTATTCTGAAACAATCTGACTACTTGAACTTGGTGCAGTGTGAGACACTTGAAGATCTAAAGCTCCATCTTCAGGGTACAGATTATGGCACTTTCCTTGCCAACGAGCCCAGCCCCCTGTCTGTATCTACTATTGATGACAAGCTTCGTGAAAAGCTTGTTATTGAATTCCAACATCTTCGGAATCACTCTGTAGAGCCTCTGTCAACATTTTTGGATTTCATTACCTACAGCTATATGATTGATAACATCATCTTGTTGATTACTGGTACATTGCACCAGAGACCTATCTCGGAGTTAATTCCAAAGTGCCATCCATTAGGCTCATTTGAGCAGATGGAAGCTATTCATGTAGCTGCAACACCAGCTGAACTTTACAATGCTGTGTTGGTGGACACACCATTGGCGCCATTCTTTGTGGATTGCATTAGCGAACAAGACTTGGATGAAATGAATATTGAGATTATCCGGAACACTCTATACAAGGCGTATTTGGAAGCTTTTTATGATTTCTGTAAGCAAATCGGAGGAACTACTGCTGATGTGATGTGCGAAATCTTAGCTTTTGAAGCCGATCGCCGTGCTATTATCATTACAATCAACTCATTTGGCACTGAACTGTCCAAGGATGACCGTGCTAAATTGTATCCCCGTTGTGGCAAACTCAACCCTGATGGTCTTGCCGCACTGGCTCGTGCTGATGATTATGAACAAGTGAAGGCTGTTGCTGAATACTATGCTGAGTATTCTGCTTTGTTTGAAGGAGCTGGCAATAATGTGGGAGACAAAACTTTGGAAGACAAATTCTTTGAACATGAGGTAAGCCTTAATGTTCATGCTTTCCTGCAACAATTCCATTTTGGAGTTTTCTATTCTTATCTGAAATTAAAGGAACAGGAGTGCCGTAACATTGTTTGGATCAGTGAATGCGTGGCCCAGAAACATCGTGCCAAAATCGACAACTACATTCCAATCTTCTAA
Protein
MKGCIFNIDAGYLEGLCRGFKCGILKQSDYLNLVQCETLEDLKLHLQGTDYGTFLANEPSPLSVSTIDDKLREKLVIEFQHLRNHSVEPLSTFLDFITYSYMIDNIILLITGTLHQRPISELIPKCHPLGSFEQMEAIHVAATPAELYNAVLVDTPLAPFFVDCISEQDLDEMNIEIIRNTLYKAYLEAFYDFCKQIGGTTADVMCEILAFEADRRAIIITINSFGTELSKDDRAKLYPRCGKLNPDGLAALARADDYEQVKAVAEYYAEYSALFEGAGNNVGDKTLEDKFFEHEVSLNVHAFLQQFHFGVFYSYLKLKEQECRNIVWISECVAQKHRAKIDNYIPIF
Summary
Description
Subunit of the integral membrane V0 complex of vacuolar ATPase. Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system.
Subunit of the integral membrane V0 complex of vacuolar ATPase. Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system. In aerobic conditions, involved in intracellular iron homeostasis, thus triggering the activity of Fe(2+) prolyl hydroxylase (PHD) enzymes, and leading to HIF1A hydroxylation and subsequent proteasomal degradation (By similarity).
Subunit of the integral membrane V0 complex of vacuolar ATPase. Vacuolar ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells, thus providing most of the energy required for transport processes in the vacuolar system. In aerobic conditions, involved in intracellular iron homeostasis, thus triggering the activity of Fe(2+) prolyl hydroxylase (PHD) enzymes, and leading to HIF1A hydroxylation and subsequent proteasomal degradation (By similarity).
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex.
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
Similarity
Belongs to the V-ATPase V0D/AC39 subunit family.
Keywords
Hydrogen ion transport
Ion transport
Transport
Complete proteome
Reference proteome
Feature
chain V-type proton ATPase subunit d
Uniprot
H9J109
Q25531
Q1HPX1
A0A2A4K6C5
I4DJR0
S4NX88
+ More
A0A194QLU4 A0A2H1VEI1 A0A212EK84 A0A0L7LPI7 A0A2R8G1V9 A0A1Y1LI32 A0A1E1WIR1 A0A2A3E9S2 V9IGC1 A0A088A285 E2BFH2 A0A026WL89 A0A0N0U3U8 A0A0C9RWV7 A0A1B0DKW9 A0A1B0C852 A0A2J7Q4F3 A0A151IM95 A0A151WV68 D6WM16 A0A1L8DFH0 A0A1B6D0F1 Q1HQS1 A0A023ERH1 K9M063 A0A1Q3FK41 A0A182K744 A0A2M3ZFP2 A0A2M4A3B9 A0A2M4BSY5 W5JQU6 A0A182Y7J9 A0A182MB12 A0A182SNI7 A0A182VBW7 A0A182W4B4 B0WSX7 A0A084VVR6 A0A182KY13 A0A182IU71 A0A182X574 Q7PRN7 A0A182R3G5 A0A182I9R2 A0A195B7F4 A0A182PPC6 A0A232FD38 K7IWG7 A0A1A9WVC7 A0A0L0C8U6 A0A1I8NZZ2 A0A336LI54 U5EUP4 E0VS26 A0A151J7H2 A0A0A1XGK0 A0A1L8EES3 T1PAZ3 A0A0K8V165 A0A034WJ70 A0A1A9YP13 A0A1A9V6L9 A0A1B0AXR7 A0A1B0A5I2 D3TMM3 A0A1J1HQJ3 A0A182NN14 A0A1W4V1L8 W8C2B5 B4Q0H4 B3MQZ0 B4NDF5 B3NSS5 A0A3B0J7G0 Q29G85 B4GW26 A0A182QXA2 A0A1L8EHA9 A0A0A9WIC5 B4MB16 B4L743 A0A0M4ELE2 B4I142 A0A0K8TPI8 Q9W4P5 A0A069DSL2 R4G8R2 A0A224XSZ4 A0A2Z1YXZ9 B4JX96 N6UJ40 A0A161M474 B4NPU6 Q5UEM9
A0A194QLU4 A0A2H1VEI1 A0A212EK84 A0A0L7LPI7 A0A2R8G1V9 A0A1Y1LI32 A0A1E1WIR1 A0A2A3E9S2 V9IGC1 A0A088A285 E2BFH2 A0A026WL89 A0A0N0U3U8 A0A0C9RWV7 A0A1B0DKW9 A0A1B0C852 A0A2J7Q4F3 A0A151IM95 A0A151WV68 D6WM16 A0A1L8DFH0 A0A1B6D0F1 Q1HQS1 A0A023ERH1 K9M063 A0A1Q3FK41 A0A182K744 A0A2M3ZFP2 A0A2M4A3B9 A0A2M4BSY5 W5JQU6 A0A182Y7J9 A0A182MB12 A0A182SNI7 A0A182VBW7 A0A182W4B4 B0WSX7 A0A084VVR6 A0A182KY13 A0A182IU71 A0A182X574 Q7PRN7 A0A182R3G5 A0A182I9R2 A0A195B7F4 A0A182PPC6 A0A232FD38 K7IWG7 A0A1A9WVC7 A0A0L0C8U6 A0A1I8NZZ2 A0A336LI54 U5EUP4 E0VS26 A0A151J7H2 A0A0A1XGK0 A0A1L8EES3 T1PAZ3 A0A0K8V165 A0A034WJ70 A0A1A9YP13 A0A1A9V6L9 A0A1B0AXR7 A0A1B0A5I2 D3TMM3 A0A1J1HQJ3 A0A182NN14 A0A1W4V1L8 W8C2B5 B4Q0H4 B3MQZ0 B4NDF5 B3NSS5 A0A3B0J7G0 Q29G85 B4GW26 A0A182QXA2 A0A1L8EHA9 A0A0A9WIC5 B4MB16 B4L743 A0A0M4ELE2 B4I142 A0A0K8TPI8 Q9W4P5 A0A069DSL2 R4G8R2 A0A224XSZ4 A0A2Z1YXZ9 B4JX96 N6UJ40 A0A161M474 B4NPU6 Q5UEM9
Pubmed
19121390
9271213
22651552
26354079
23622113
22118469
+ More
26227816 28004739 20798317 24508170 30249741 18362917 19820115 17204158 17510324 24945155 26483478 20920257 23761445 25244985 24438588 20966253 12364791 14747013 17210077 28648823 20075255 26108605 20566863 25830018 25315136 25348373 20353571 24495485 17994087 17550304 15632085 25401762 26369729 10731132 12537572 12537569 26334808 27558665 23537049
26227816 28004739 20798317 24508170 30249741 18362917 19820115 17204158 17510324 24945155 26483478 20920257 23761445 25244985 24438588 20966253 12364791 14747013 17210077 28648823 20075255 26108605 20566863 25830018 25315136 25348373 20353571 24495485 17994087 17550304 15632085 25401762 26369729 10731132 12537572 12537569 26334808 27558665 23537049
EMBL
BABH01010534
X98825
DQ443281
ABF51370.1
NWSH01000081
PCG79815.1
+ More
AK401528 KQ459601 BAM18150.1 KPI93738.1 GAIX01009169 JAA83391.1 KQ461196 KPJ06497.1 ODYU01001874 SOQ38684.1 AGBW02014319 OWR41875.1 JTDY01000385 KOB77433.1 KY921847 AVP73919.1 GEZM01056946 JAV72551.1 GDQN01004151 JAT86903.1 KZ288311 PBC28477.1 JR043001 AEY59707.1 GL448037 EFN85494.1 KK107154 QOIP01000001 EZA56805.1 RLU27159.1 KQ435881 KOX69852.1 GBYB01012335 GBYB01012336 JAG82102.1 JAG82103.1 AJVK01006516 AJVK01006517 AJWK01000173 NEVH01018383 PNF23460.1 KQ977063 KYN05993.1 KQ982706 KYQ51830.1 KQ971343 EFA03371.1 GFDF01008974 JAV05110.1 GEDC01018161 JAS19137.1 DQ440373 CH477719 ABF18406.1 EAT36933.1 JXUM01016045 JXUM01108598 GAPW01002337 KQ565246 KQ560448 JAC11261.1 KXJ71185.1 KXJ82372.1 JN676087 AFQ00928.1 GFDL01007098 JAV27947.1 GGFM01006550 MBW27301.1 GGFK01001920 MBW35241.1 GGFJ01006962 MBW56103.1 ADMH02000604 ETN65678.1 AXCM01004867 DS232077 EDS34105.1 ATLV01017216 KE525157 KFB42060.1 AXCP01004934 AAAB01008847 EAA06911.5 APCN01001027 KQ976565 KYM80441.1 NNAY01000440 OXU28379.1 JRES01000755 KNC28657.1 UFQS01000015 UFQT01000015 SSW97348.1 SSX17734.1 GANO01001377 JAB58494.1 DS235742 EEB16182.1 KQ979679 KYN19851.1 GBXI01004664 JAD09628.1 GFDG01001601 JAV17198.1 KA645902 AFP60531.1 GDHF01019718 JAI32596.1 GAKP01005124 JAC53828.1 JXJN01005350 CCAG010014759 EZ422675 ADD18951.1 CVRI01000014 CRK89802.1 GAMC01003197 JAC03359.1 CM000162 EDX01258.1 CH902622 EDV34195.1 CH964239 EDW82861.1 CH954180 EDV45755.1 OUUW01000003 SPP78184.1 CH379064 EAL32225.1 CH479193 EDW26871.1 AXCN02001590 GFDG01000755 JAV18044.1 GBHO01036458 GBRD01016805 JAG07146.1 JAG49022.1 CH940655 EDW66425.1 CH933812 EDW06189.1 CP012528 ALC48234.1 CH480819 EDW53223.1 GDAI01001540 JAI16063.1 AE014298 AY069530 GBGD01001994 JAC86895.1 ACPB03001482 GAHY01000527 JAA76983.1 GFTR01005337 JAW11089.1 KU665627 AOM63315.1 CH916376 EDV95372.1 APGK01018036 KB740064 KB632132 ENN81765.1 ERL89014.1 GEMB01004298 JAR98974.1 CH964291 EDW86171.1 AY750873 AAV31420.1
AK401528 KQ459601 BAM18150.1 KPI93738.1 GAIX01009169 JAA83391.1 KQ461196 KPJ06497.1 ODYU01001874 SOQ38684.1 AGBW02014319 OWR41875.1 JTDY01000385 KOB77433.1 KY921847 AVP73919.1 GEZM01056946 JAV72551.1 GDQN01004151 JAT86903.1 KZ288311 PBC28477.1 JR043001 AEY59707.1 GL448037 EFN85494.1 KK107154 QOIP01000001 EZA56805.1 RLU27159.1 KQ435881 KOX69852.1 GBYB01012335 GBYB01012336 JAG82102.1 JAG82103.1 AJVK01006516 AJVK01006517 AJWK01000173 NEVH01018383 PNF23460.1 KQ977063 KYN05993.1 KQ982706 KYQ51830.1 KQ971343 EFA03371.1 GFDF01008974 JAV05110.1 GEDC01018161 JAS19137.1 DQ440373 CH477719 ABF18406.1 EAT36933.1 JXUM01016045 JXUM01108598 GAPW01002337 KQ565246 KQ560448 JAC11261.1 KXJ71185.1 KXJ82372.1 JN676087 AFQ00928.1 GFDL01007098 JAV27947.1 GGFM01006550 MBW27301.1 GGFK01001920 MBW35241.1 GGFJ01006962 MBW56103.1 ADMH02000604 ETN65678.1 AXCM01004867 DS232077 EDS34105.1 ATLV01017216 KE525157 KFB42060.1 AXCP01004934 AAAB01008847 EAA06911.5 APCN01001027 KQ976565 KYM80441.1 NNAY01000440 OXU28379.1 JRES01000755 KNC28657.1 UFQS01000015 UFQT01000015 SSW97348.1 SSX17734.1 GANO01001377 JAB58494.1 DS235742 EEB16182.1 KQ979679 KYN19851.1 GBXI01004664 JAD09628.1 GFDG01001601 JAV17198.1 KA645902 AFP60531.1 GDHF01019718 JAI32596.1 GAKP01005124 JAC53828.1 JXJN01005350 CCAG010014759 EZ422675 ADD18951.1 CVRI01000014 CRK89802.1 GAMC01003197 JAC03359.1 CM000162 EDX01258.1 CH902622 EDV34195.1 CH964239 EDW82861.1 CH954180 EDV45755.1 OUUW01000003 SPP78184.1 CH379064 EAL32225.1 CH479193 EDW26871.1 AXCN02001590 GFDG01000755 JAV18044.1 GBHO01036458 GBRD01016805 JAG07146.1 JAG49022.1 CH940655 EDW66425.1 CH933812 EDW06189.1 CP012528 ALC48234.1 CH480819 EDW53223.1 GDAI01001540 JAI16063.1 AE014298 AY069530 GBGD01001994 JAC86895.1 ACPB03001482 GAHY01000527 JAA76983.1 GFTR01005337 JAW11089.1 KU665627 AOM63315.1 CH916376 EDV95372.1 APGK01018036 KB740064 KB632132 ENN81765.1 ERL89014.1 GEMB01004298 JAR98974.1 CH964291 EDW86171.1 AY750873 AAV31420.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000242457 UP000005203 UP000008237 UP000053097 UP000279307 UP000053105 UP000092462 UP000092461 UP000235965 UP000078542 UP000075809 UP000007266 UP000008820 UP000069940 UP000249989 UP000075881 UP000000673 UP000076408 UP000075883 UP000075901 UP000075903 UP000075920 UP000002320 UP000030765 UP000075882 UP000075880 UP000076407 UP000007062 UP000075900 UP000075840 UP000078540 UP000075885 UP000215335 UP000002358 UP000091820 UP000037069 UP000095300 UP000009046 UP000078492 UP000095301 UP000092443 UP000078200 UP000092460 UP000092445 UP000092444 UP000183832 UP000075884 UP000192221 UP000002282 UP000007801 UP000007798 UP000008711 UP000268350 UP000001819 UP000008744 UP000075886 UP000008792 UP000009192 UP000092553 UP000001292 UP000000803 UP000015103 UP000001070 UP000019118 UP000030742
UP000242457 UP000005203 UP000008237 UP000053097 UP000279307 UP000053105 UP000092462 UP000092461 UP000235965 UP000078542 UP000075809 UP000007266 UP000008820 UP000069940 UP000249989 UP000075881 UP000000673 UP000076408 UP000075883 UP000075901 UP000075903 UP000075920 UP000002320 UP000030765 UP000075882 UP000075880 UP000076407 UP000007062 UP000075900 UP000075840 UP000078540 UP000075885 UP000215335 UP000002358 UP000091820 UP000037069 UP000095300 UP000009046 UP000078492 UP000095301 UP000092443 UP000078200 UP000092460 UP000092445 UP000092444 UP000183832 UP000075884 UP000192221 UP000002282 UP000007801 UP000007798 UP000008711 UP000268350 UP000001819 UP000008744 UP000075886 UP000008792 UP000009192 UP000092553 UP000001292 UP000000803 UP000015103 UP000001070 UP000019118 UP000030742
Pfam
PF01992 vATP-synt_AC39
Interpro
SUPFAM
SSF103486
SSF103486
Gene 3D
ProteinModelPortal
H9J109
Q25531
Q1HPX1
A0A2A4K6C5
I4DJR0
S4NX88
+ More
A0A194QLU4 A0A2H1VEI1 A0A212EK84 A0A0L7LPI7 A0A2R8G1V9 A0A1Y1LI32 A0A1E1WIR1 A0A2A3E9S2 V9IGC1 A0A088A285 E2BFH2 A0A026WL89 A0A0N0U3U8 A0A0C9RWV7 A0A1B0DKW9 A0A1B0C852 A0A2J7Q4F3 A0A151IM95 A0A151WV68 D6WM16 A0A1L8DFH0 A0A1B6D0F1 Q1HQS1 A0A023ERH1 K9M063 A0A1Q3FK41 A0A182K744 A0A2M3ZFP2 A0A2M4A3B9 A0A2M4BSY5 W5JQU6 A0A182Y7J9 A0A182MB12 A0A182SNI7 A0A182VBW7 A0A182W4B4 B0WSX7 A0A084VVR6 A0A182KY13 A0A182IU71 A0A182X574 Q7PRN7 A0A182R3G5 A0A182I9R2 A0A195B7F4 A0A182PPC6 A0A232FD38 K7IWG7 A0A1A9WVC7 A0A0L0C8U6 A0A1I8NZZ2 A0A336LI54 U5EUP4 E0VS26 A0A151J7H2 A0A0A1XGK0 A0A1L8EES3 T1PAZ3 A0A0K8V165 A0A034WJ70 A0A1A9YP13 A0A1A9V6L9 A0A1B0AXR7 A0A1B0A5I2 D3TMM3 A0A1J1HQJ3 A0A182NN14 A0A1W4V1L8 W8C2B5 B4Q0H4 B3MQZ0 B4NDF5 B3NSS5 A0A3B0J7G0 Q29G85 B4GW26 A0A182QXA2 A0A1L8EHA9 A0A0A9WIC5 B4MB16 B4L743 A0A0M4ELE2 B4I142 A0A0K8TPI8 Q9W4P5 A0A069DSL2 R4G8R2 A0A224XSZ4 A0A2Z1YXZ9 B4JX96 N6UJ40 A0A161M474 B4NPU6 Q5UEM9
A0A194QLU4 A0A2H1VEI1 A0A212EK84 A0A0L7LPI7 A0A2R8G1V9 A0A1Y1LI32 A0A1E1WIR1 A0A2A3E9S2 V9IGC1 A0A088A285 E2BFH2 A0A026WL89 A0A0N0U3U8 A0A0C9RWV7 A0A1B0DKW9 A0A1B0C852 A0A2J7Q4F3 A0A151IM95 A0A151WV68 D6WM16 A0A1L8DFH0 A0A1B6D0F1 Q1HQS1 A0A023ERH1 K9M063 A0A1Q3FK41 A0A182K744 A0A2M3ZFP2 A0A2M4A3B9 A0A2M4BSY5 W5JQU6 A0A182Y7J9 A0A182MB12 A0A182SNI7 A0A182VBW7 A0A182W4B4 B0WSX7 A0A084VVR6 A0A182KY13 A0A182IU71 A0A182X574 Q7PRN7 A0A182R3G5 A0A182I9R2 A0A195B7F4 A0A182PPC6 A0A232FD38 K7IWG7 A0A1A9WVC7 A0A0L0C8U6 A0A1I8NZZ2 A0A336LI54 U5EUP4 E0VS26 A0A151J7H2 A0A0A1XGK0 A0A1L8EES3 T1PAZ3 A0A0K8V165 A0A034WJ70 A0A1A9YP13 A0A1A9V6L9 A0A1B0AXR7 A0A1B0A5I2 D3TMM3 A0A1J1HQJ3 A0A182NN14 A0A1W4V1L8 W8C2B5 B4Q0H4 B3MQZ0 B4NDF5 B3NSS5 A0A3B0J7G0 Q29G85 B4GW26 A0A182QXA2 A0A1L8EHA9 A0A0A9WIC5 B4MB16 B4L743 A0A0M4ELE2 B4I142 A0A0K8TPI8 Q9W4P5 A0A069DSL2 R4G8R2 A0A224XSZ4 A0A2Z1YXZ9 B4JX96 N6UJ40 A0A161M474 B4NPU6 Q5UEM9
PDB
6O7X
E-value=8.82683e-85,
Score=798
Ontologies
KEGG
PATHWAY
GO
PANTHER
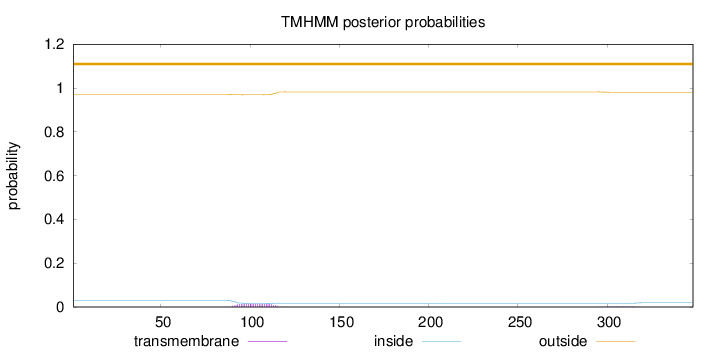
Topology
Length:
348
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.357740000000001
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.02975
outside
1 - 348
Population Genetic Test Statistics
Pi
188.901717
Theta
160.816885
Tajima's D
0.549177
CLR
1.188842
CSRT
0.529923503824809
Interpretation
Uncertain
