Gene
KWMTBOMO02061 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011168
Annotation
S-adenosyl-L-homocysteine_hydrolase_[Bombyx_mori]
Full name
Adenosylhomocysteinase
Alternative Name
S-adenosyl-L-homocysteine hydrolase
Location in the cell
Cytoplasmic Reliability : 3.987
Sequence
CDS
ATGAAGCCTCCTTACAAAATCGCTGACGAGAAATTGGCTGAATGGGGCCGAAAAGAAATTATGTTAGCAGAGAAAGAAATGCCAGGCCTTATGGCATGTCGTCGGAAATATGCTCCAGCTAAAATACTCAAAGGTGCCAGAATAGCCGGCAGCTTACACATGACAGTACAAACAGCGGTTCTTATTGAGACTTTAATAGAATTGGGAGCAGAGGTACAATGGTCCAGCAGTAACATTTATAGTACACAAGACGAGGCCGCAGCTGCACTGGTTGCCGTAGGAATACCCATCTATGCCTGGAAGGGAGAAACTGATGACGAGTATATTTGGTGTATTGAACAAACATTGATCTTCCCTGATGGAAAGCCTCTCAATATGATCTTGGATGATGGGGGTGACTTAACAAACTTAGTCCACACTAAGTACCCCGATCTTTTAAAAGACGTAAAAGGTATCACTGAAGAAACTACAACCGGGGTACATAATTTGTATAAAATGTTCCGAGAAGGACTTTTAAAAGTTCCTGCAATCAACGTTAACGATTCAGTAACAAAAAGCAAATTCGACAACTTGTATGGATGTAGGGAGTCTTTGCTCGACGGAATCAAAAGGGCAACAGACATAATGATTGCCGGGAAAGTTTGTGTAGTGGCAGGTTATGGTGACGTTGGAAAAGGATGCGCCCAAGCGTTCAAAGGTTTTGGCGGTAGAGTGATCGTTACCGAAATCGATCCCATCAACGCACTTCAGGCCGCAATGGAAGGTTTTCAAGTGACTACTATGGAGGAAGCCGCTGAAGTGGGTCAAATCTTTGTCACCACGACGGGAAATATTGACATTATATGCAAAGAACATCTTCTTAGAATGAAGGACGATGCAATTGTGTGCAACATTGGTCACTTTGATTGCGAAATTGATGTTGCATGGCTAGATAACAACGCAAAAAAAGTTAACATCAAACAACATGTTGATCGTTATGAATTAGAAAATGGTAACCATATTATTGTTCTTGCTGCAGGAAGACTGGTTAATTTAGGTTGCGCAACCGGCCATTCCTCTTTTGTAATGTCTAATTCATTCACAAATCAAGTTCTAGCTCAGATAGAGCTGTGGACCAAAGCAGACACATATCCAATAGGTGTCCACACACTACCTAAGAAACTTGACGAGGAAGTAGCTGCCTTGCATTTGGATCACCTTGGTGTAAAACTGACGAAGTTAACACCTAAACAAGCCAAATACATTGGTGTCTCTCGAGAAGGACCTTACAAGCCAGATCATTACAGATATTAA
Protein
MKPPYKIADEKLAEWGRKEIMLAEKEMPGLMACRRKYAPAKILKGARIAGSLHMTVQTAVLIETLIELGAEVQWSSSNIYSTQDEAAAALVAVGIPIYAWKGETDDEYIWCIEQTLIFPDGKPLNMILDDGGDLTNLVHTKYPDLLKDVKGITEETTTGVHNLYKMFREGLLKVPAINVNDSVTKSKFDNLYGCRESLLDGIKRATDIMIAGKVCVVAGYGDVGKGCAQAFKGFGGRVIVTEIDPINALQAAMEGFQVTTMEEAAEVGQIFVTTTGNIDIICKEHLLRMKDDAIVCNIGHFDCEIDVAWLDNNAKKVNIKQHVDRYELENGNHIIVLAAGRLVNLGCATGHSSFVMSNSFTNQVLAQIELWTKADTYPIGVHTLPKKLDEEVAALHLDHLGVKLTKLTPKQAKYIGVSREGPYKPDHYRY
Summary
Description
Adenosylhomocysteine is a competitive inhibitor of S-adenosyl-L-methionine-dependent methyl transferase reactions; therefore adenosylhomocysteinase may play a key role in the control of methylations via regulation of the intracellular concentration of adenosylhomocysteine.
Catalytic Activity
H2O + S-adenosyl-L-homocysteine = adenosine + L-homocysteine
Cofactor
NAD(+)
Subunit
Interacts with AhcyL1; the interaction may negatively regulate Ahcy catalytic activity.
Similarity
Belongs to the adenosylhomocysteinase family.
Keywords
Complete proteome
Hydrolase
NAD
One-carbon metabolism
Reference proteome
Feature
chain Adenosylhomocysteinase
Uniprot
Q1HPU7
A0A212EK57
A0A2A4K6D7
A0A2H1VCY7
S4PFC2
A0A194QSI2
+ More
A0JCJ9 I4DJ26 A0A220K8Q1 A0A2A4K821 A0A0L7LPR8 A0A222AJ25 H9JNR4 A0A182QB80 A0A0J7L559 A0A2M4BNW7 A0A0H4J5A1 A0A182N1X4 D6WMI3 A0A2M3ZH49 A0A182FH88 A0A182PPC4 A0A232F0W8 V5GRK7 E2A3R3 A0A2M4A5U0 J3JZ28 T1DMZ8 A0A1B6G340 A0A182W4B6 A0A182T5F0 A0A182MEY8 A0A182Y7K2 A0A182IJX3 A0A1B6MRU4 Q1HQR0 A0A023ES51 A0A182MZP9 A0A084VT28 T1PCH0 A0A182L6A9 O76757 A0A182X572 A0A182VBX5 A0A182I9R4 A0A182U980 A0A2A3EM47 A0A1L8EDS3 A0A1I8NVD1 A0A026WTZ2 A0A0L0C832 A0A088A1B6 A0A1Q3FVZ4 A0A182R3G7 A0A2M4CTK5 A0A158NKA3 U5EZ85 F4W8K2 A0A1J1HTJ9 A0A182IW45 A0A2M4CUT3 A0A310SGQ7 A0A084VVR9 B3MR98 A0A1L8DZN4 B4MSP6 B3NTL4 A0A1W4VH79 B4M6U8 B4L1T2 B4PWM7 E2BIK4 E9IBV6 A0A3B0J841 Q29G51 A0A0A1XL48 B4GVX7 A0A1Y1MK78 A0A0M5JDP9 B4JLP1 A0A1W4XAP6 X2JF40 Q27580 B4R576 A0A034VRE6 B4ILS9 A0A182R4F8 A0A2J7Q4G0 A0A1B6D2V7 A0A151WIQ5 A0A0K8U908 A0A067QR94 A0A336K2G9 A0A195BAW4 A0A195EHN6 W8CD81 A0A0L7R5T9 A0A069DUL7 A0A224XQH0 A0A161ME74
A0JCJ9 I4DJ26 A0A220K8Q1 A0A2A4K821 A0A0L7LPR8 A0A222AJ25 H9JNR4 A0A182QB80 A0A0J7L559 A0A2M4BNW7 A0A0H4J5A1 A0A182N1X4 D6WMI3 A0A2M3ZH49 A0A182FH88 A0A182PPC4 A0A232F0W8 V5GRK7 E2A3R3 A0A2M4A5U0 J3JZ28 T1DMZ8 A0A1B6G340 A0A182W4B6 A0A182T5F0 A0A182MEY8 A0A182Y7K2 A0A182IJX3 A0A1B6MRU4 Q1HQR0 A0A023ES51 A0A182MZP9 A0A084VT28 T1PCH0 A0A182L6A9 O76757 A0A182X572 A0A182VBX5 A0A182I9R4 A0A182U980 A0A2A3EM47 A0A1L8EDS3 A0A1I8NVD1 A0A026WTZ2 A0A0L0C832 A0A088A1B6 A0A1Q3FVZ4 A0A182R3G7 A0A2M4CTK5 A0A158NKA3 U5EZ85 F4W8K2 A0A1J1HTJ9 A0A182IW45 A0A2M4CUT3 A0A310SGQ7 A0A084VVR9 B3MR98 A0A1L8DZN4 B4MSP6 B3NTL4 A0A1W4VH79 B4M6U8 B4L1T2 B4PWM7 E2BIK4 E9IBV6 A0A3B0J841 Q29G51 A0A0A1XL48 B4GVX7 A0A1Y1MK78 A0A0M5JDP9 B4JLP1 A0A1W4XAP6 X2JF40 Q27580 B4R576 A0A034VRE6 B4ILS9 A0A182R4F8 A0A2J7Q4G0 A0A1B6D2V7 A0A151WIQ5 A0A0K8U908 A0A067QR94 A0A336K2G9 A0A195BAW4 A0A195EHN6 W8CD81 A0A0L7R5T9 A0A069DUL7 A0A224XQH0 A0A161ME74
EC Number
3.3.1.1
Pubmed
22118469
23622113
26354079
22651552
28630352
26227816
+ More
19121390 18362917 19820115 28648823 20798317 22516182 25244985 17204158 17510324 24945155 26483478 24438588 25315136 20966253 12364791 24508170 30249741 26108605 21347285 21719571 17994087 17550304 21282665 15632085 25830018 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9037110 12537569 27313316 25348373 24845553 24495485 26334808
19121390 18362917 19820115 28648823 20798317 22516182 25244985 17204158 17510324 24945155 26483478 24438588 25315136 20966253 12364791 24508170 30249741 26108605 21347285 21719571 17994087 17550304 21282665 15632085 25830018 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9037110 12537569 27313316 25348373 24845553 24495485 26334808
EMBL
DQ443305
ABF51394.1
AGBW02014319
OWR41879.1
NWSH01000081
PCG79801.1
+ More
ODYU01001874 SOQ38687.1 GAIX01004082 JAA88478.1 KQ461196 KPJ06501.1 AB282637 BAF36817.1 AK401294 BAM17916.1 MF319733 ASJ26447.1 PCG79800.1 JTDY01000385 KOB77432.1 KY563910 ASO76366.1 BABH01042252 AXCN02000974 LBMM01000555 KMQ98067.1 GGFJ01005522 MBW54663.1 KP689345 AKO63327.1 KQ971343 EFA04274.1 GGFM01007039 MBW27790.1 NNAY01001377 OXU24183.1 GALX01004209 JAB64257.1 GL436457 EFN71955.1 GGFK01002761 MBW36082.1 BT128509 AEE63466.1 GAMD01003295 JAA98295.1 GECZ01012911 JAS56858.1 AXCM01002930 GEBQ01001310 JAT38667.1 DQ440384 CH477503 ABF18417.1 EAT39891.1 JXUM01075535 GAPW01001551 KQ562892 JAC12047.1 KXJ74898.1 ATLV01016154 KE525057 KFB41122.1 KA645830 AFP60459.1 AF080546 AAAB01008847 APCN01001029 KZ288215 PBC32760.1 GFDG01001931 JAV16868.1 KK107105 QOIP01000011 EZA59535.1 RLU17141.1 JRES01000777 KNC28405.1 GFDL01003265 JAV31780.1 GGFL01004472 MBW68650.1 ADTU01018755 GANO01001248 JAB58623.1 GL887908 EGI69493.1 CVRI01000020 CRK90708.1 GGFL01004420 MBW68598.1 KQ764881 OAD54323.1 ATLV01017216 KE525157 KFB42063.1 CH902622 EDV34303.1 GFDF01002188 JAV11896.1 CH963851 EDW75135.1 CH954180 EDV47018.1 CH940653 EDW62515.1 CH933810 EDW06735.2 CM000162 EDX01773.1 GL448516 EFN84451.1 GL762137 EFZ21986.1 OUUW01000003 SPP78317.1 CH379064 EAL32259.1 GBXI01003029 JAD11263.1 CH479193 EDW26822.1 GEZM01032229 JAV84755.1 CP012528 ALC49118.1 CH916371 EDV91652.1 AE014298 AHN59738.1 X95636 AY102668 CM000366 EDX17977.1 GAKP01014829 GAKP01014826 JAC44123.1 CH480888 EDW55379.1 NEVH01018383 PNF23459.1 GEDC01017322 JAS19976.1 KQ983082 KYQ47728.1 GDHF01029504 GDHF01009913 JAI22810.1 JAI42401.1 KK853038 KDR12193.1 UFQS01000063 UFQT01000063 SSW98879.1 SSX19265.1 KQ976537 KYM81334.1 KQ978881 KYN27773.1 GAMC01002406 JAC04150.1 KQ414652 KOC66131.1 GBGD01001527 JAC87362.1 GFTR01006185 JAW10241.1 GEMB01000308 JAS02811.1
ODYU01001874 SOQ38687.1 GAIX01004082 JAA88478.1 KQ461196 KPJ06501.1 AB282637 BAF36817.1 AK401294 BAM17916.1 MF319733 ASJ26447.1 PCG79800.1 JTDY01000385 KOB77432.1 KY563910 ASO76366.1 BABH01042252 AXCN02000974 LBMM01000555 KMQ98067.1 GGFJ01005522 MBW54663.1 KP689345 AKO63327.1 KQ971343 EFA04274.1 GGFM01007039 MBW27790.1 NNAY01001377 OXU24183.1 GALX01004209 JAB64257.1 GL436457 EFN71955.1 GGFK01002761 MBW36082.1 BT128509 AEE63466.1 GAMD01003295 JAA98295.1 GECZ01012911 JAS56858.1 AXCM01002930 GEBQ01001310 JAT38667.1 DQ440384 CH477503 ABF18417.1 EAT39891.1 JXUM01075535 GAPW01001551 KQ562892 JAC12047.1 KXJ74898.1 ATLV01016154 KE525057 KFB41122.1 KA645830 AFP60459.1 AF080546 AAAB01008847 APCN01001029 KZ288215 PBC32760.1 GFDG01001931 JAV16868.1 KK107105 QOIP01000011 EZA59535.1 RLU17141.1 JRES01000777 KNC28405.1 GFDL01003265 JAV31780.1 GGFL01004472 MBW68650.1 ADTU01018755 GANO01001248 JAB58623.1 GL887908 EGI69493.1 CVRI01000020 CRK90708.1 GGFL01004420 MBW68598.1 KQ764881 OAD54323.1 ATLV01017216 KE525157 KFB42063.1 CH902622 EDV34303.1 GFDF01002188 JAV11896.1 CH963851 EDW75135.1 CH954180 EDV47018.1 CH940653 EDW62515.1 CH933810 EDW06735.2 CM000162 EDX01773.1 GL448516 EFN84451.1 GL762137 EFZ21986.1 OUUW01000003 SPP78317.1 CH379064 EAL32259.1 GBXI01003029 JAD11263.1 CH479193 EDW26822.1 GEZM01032229 JAV84755.1 CP012528 ALC49118.1 CH916371 EDV91652.1 AE014298 AHN59738.1 X95636 AY102668 CM000366 EDX17977.1 GAKP01014829 GAKP01014826 JAC44123.1 CH480888 EDW55379.1 NEVH01018383 PNF23459.1 GEDC01017322 JAS19976.1 KQ983082 KYQ47728.1 GDHF01029504 GDHF01009913 JAI22810.1 JAI42401.1 KK853038 KDR12193.1 UFQS01000063 UFQT01000063 SSW98879.1 SSX19265.1 KQ976537 KYM81334.1 KQ978881 KYN27773.1 GAMC01002406 JAC04150.1 KQ414652 KOC66131.1 GBGD01001527 JAC87362.1 GFTR01006185 JAW10241.1 GEMB01000308 JAS02811.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000037510
UP000005204
UP000075886
+ More
UP000036403 UP000075884 UP000007266 UP000069272 UP000075885 UP000215335 UP000000311 UP000075920 UP000075901 UP000075883 UP000076408 UP000075880 UP000008820 UP000069940 UP000249989 UP000030765 UP000095301 UP000075882 UP000007062 UP000076407 UP000075903 UP000075840 UP000075902 UP000242457 UP000095300 UP000053097 UP000279307 UP000037069 UP000005203 UP000075900 UP000005205 UP000007755 UP000183832 UP000007801 UP000007798 UP000008711 UP000192221 UP000008792 UP000009192 UP000002282 UP000008237 UP000268350 UP000001819 UP000008744 UP000092553 UP000001070 UP000192223 UP000000803 UP000000304 UP000001292 UP000235965 UP000075809 UP000027135 UP000078540 UP000078492 UP000053825
UP000036403 UP000075884 UP000007266 UP000069272 UP000075885 UP000215335 UP000000311 UP000075920 UP000075901 UP000075883 UP000076408 UP000075880 UP000008820 UP000069940 UP000249989 UP000030765 UP000095301 UP000075882 UP000007062 UP000076407 UP000075903 UP000075840 UP000075902 UP000242457 UP000095300 UP000053097 UP000279307 UP000037069 UP000005203 UP000075900 UP000005205 UP000007755 UP000183832 UP000007801 UP000007798 UP000008711 UP000192221 UP000008792 UP000009192 UP000002282 UP000008237 UP000268350 UP000001819 UP000008744 UP000092553 UP000001070 UP000192223 UP000000803 UP000000304 UP000001292 UP000235965 UP000075809 UP000027135 UP000078540 UP000078492 UP000053825
Interpro
IPR000043
Adenosylhomocysteinase-like
+ More
IPR015878 Ado_hCys_hydrolase_NAD-bd
IPR034373 Adenosylhomocysteinase
IPR042172 Adenosylhomocyst_ase-like_sf
IPR036291 NAD(P)-bd_dom_sf
IPR020082 S-Ado-L-homoCys_hydrolase_CS
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR030470 UbiA_prenylTrfase_CS
IPR000537 UbiA_prenyltransferase
IPR006369 Protohaem_IX_farnesylTrfase
IPR015878 Ado_hCys_hydrolase_NAD-bd
IPR034373 Adenosylhomocysteinase
IPR042172 Adenosylhomocyst_ase-like_sf
IPR036291 NAD(P)-bd_dom_sf
IPR020082 S-Ado-L-homoCys_hydrolase_CS
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR030470 UbiA_prenylTrfase_CS
IPR000537 UbiA_prenyltransferase
IPR006369 Protohaem_IX_farnesylTrfase
SUPFAM
SSF51735
SSF51735
Gene 3D
ProteinModelPortal
Q1HPU7
A0A212EK57
A0A2A4K6D7
A0A2H1VCY7
S4PFC2
A0A194QSI2
+ More
A0JCJ9 I4DJ26 A0A220K8Q1 A0A2A4K821 A0A0L7LPR8 A0A222AJ25 H9JNR4 A0A182QB80 A0A0J7L559 A0A2M4BNW7 A0A0H4J5A1 A0A182N1X4 D6WMI3 A0A2M3ZH49 A0A182FH88 A0A182PPC4 A0A232F0W8 V5GRK7 E2A3R3 A0A2M4A5U0 J3JZ28 T1DMZ8 A0A1B6G340 A0A182W4B6 A0A182T5F0 A0A182MEY8 A0A182Y7K2 A0A182IJX3 A0A1B6MRU4 Q1HQR0 A0A023ES51 A0A182MZP9 A0A084VT28 T1PCH0 A0A182L6A9 O76757 A0A182X572 A0A182VBX5 A0A182I9R4 A0A182U980 A0A2A3EM47 A0A1L8EDS3 A0A1I8NVD1 A0A026WTZ2 A0A0L0C832 A0A088A1B6 A0A1Q3FVZ4 A0A182R3G7 A0A2M4CTK5 A0A158NKA3 U5EZ85 F4W8K2 A0A1J1HTJ9 A0A182IW45 A0A2M4CUT3 A0A310SGQ7 A0A084VVR9 B3MR98 A0A1L8DZN4 B4MSP6 B3NTL4 A0A1W4VH79 B4M6U8 B4L1T2 B4PWM7 E2BIK4 E9IBV6 A0A3B0J841 Q29G51 A0A0A1XL48 B4GVX7 A0A1Y1MK78 A0A0M5JDP9 B4JLP1 A0A1W4XAP6 X2JF40 Q27580 B4R576 A0A034VRE6 B4ILS9 A0A182R4F8 A0A2J7Q4G0 A0A1B6D2V7 A0A151WIQ5 A0A0K8U908 A0A067QR94 A0A336K2G9 A0A195BAW4 A0A195EHN6 W8CD81 A0A0L7R5T9 A0A069DUL7 A0A224XQH0 A0A161ME74
A0JCJ9 I4DJ26 A0A220K8Q1 A0A2A4K821 A0A0L7LPR8 A0A222AJ25 H9JNR4 A0A182QB80 A0A0J7L559 A0A2M4BNW7 A0A0H4J5A1 A0A182N1X4 D6WMI3 A0A2M3ZH49 A0A182FH88 A0A182PPC4 A0A232F0W8 V5GRK7 E2A3R3 A0A2M4A5U0 J3JZ28 T1DMZ8 A0A1B6G340 A0A182W4B6 A0A182T5F0 A0A182MEY8 A0A182Y7K2 A0A182IJX3 A0A1B6MRU4 Q1HQR0 A0A023ES51 A0A182MZP9 A0A084VT28 T1PCH0 A0A182L6A9 O76757 A0A182X572 A0A182VBX5 A0A182I9R4 A0A182U980 A0A2A3EM47 A0A1L8EDS3 A0A1I8NVD1 A0A026WTZ2 A0A0L0C832 A0A088A1B6 A0A1Q3FVZ4 A0A182R3G7 A0A2M4CTK5 A0A158NKA3 U5EZ85 F4W8K2 A0A1J1HTJ9 A0A182IW45 A0A2M4CUT3 A0A310SGQ7 A0A084VVR9 B3MR98 A0A1L8DZN4 B4MSP6 B3NTL4 A0A1W4VH79 B4M6U8 B4L1T2 B4PWM7 E2BIK4 E9IBV6 A0A3B0J841 Q29G51 A0A0A1XL48 B4GVX7 A0A1Y1MK78 A0A0M5JDP9 B4JLP1 A0A1W4XAP6 X2JF40 Q27580 B4R576 A0A034VRE6 B4ILS9 A0A182R4F8 A0A2J7Q4G0 A0A1B6D2V7 A0A151WIQ5 A0A0K8U908 A0A067QR94 A0A336K2G9 A0A195BAW4 A0A195EHN6 W8CD81 A0A0L7R5T9 A0A069DUL7 A0A224XQH0 A0A161ME74
PDB
5W4B
E-value=0,
Score=1748
Ontologies
KEGG
PATHWAY
GO
PANTHER
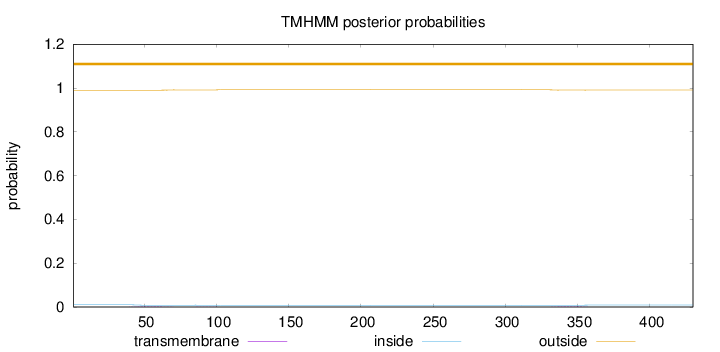
Topology
Length:
430
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0768100000000001
Exp number, first 60 AAs:
0.03202
Total prob of N-in:
0.01034
outside
1 - 430
Population Genetic Test Statistics
Pi
173.558112
Theta
171.997036
Tajima's D
0.187725
CLR
0.393889
CSRT
0.425778711064447
Interpretation
Uncertain
