Gene
KWMTBOMO02060 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002972
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_importin_subunit_beta-1_[Bombyx_mori]
Location in the cell
Golgi Reliability : 2.742
Sequence
CDS
ATGCACGCAGAAACAACATTAACGCTTATACAAATACTTGAGAAGACAGTTTCACCTGATCGAAATGAATTGGAAGCAGCAGTTAGATACCTCGACCATGCTGCCACCACAAATTTCACAACATTCATCAAAATGTTGTCCGATGTCCTTCTCCAAGGTGGTAACAGTCAAGTGGCTAGGATGGCAGCTGGTTTGCAATTGAAAAATCATCTCACTTCTAAAGATCCTACACTTAAACAACAATATCAACAAAGATGGCTGGCTTTGGCAGAAGATGTTCGACTTTATATAAAAGAAAATATATTAGCTGCTATAGGAACAGAGAATAGTCGTCCAAGTTCAGCAGCACAGTGTGTGGCGTATGTGGCAGTAGCTGAGCTCCCAGTAGGACAATGGAATGATCTAATACCAATTCTTGTAGAAAATGTTGTTCATGTCCAATCTACTGAACTCAAAAAGGAAGCGACTTTGGAGGCTATTGGCTACATTTGTCAGGATATTGATGCTGAAGTATTAACAGAAAGAAGTAATCAAATTCTTACGGCCATCATACATGGCATGAGGTCAACTGAACCCAGTAATCATGTTCGACTAGCTGCTACTCAAGCTTTGCTCAACTCTCTAGAATTCACCAGAGCGAACTTTGAAAAAGAAAATGAAAGAAACTTTATTATGGAGGTTGTCTGTGAAGCCACACAGTCTAGTGACATGAGAATCAGCGTTGCTGCTTTGCAATGCTTAGTGAAAATTTTATCACTTTATTATCAACATATGGAACCTTACATGGGACAAGCTCTATTCCCCATAACACTTGAGGCTATGAAATCTGATATTGATGAGATTTCATTACAAGGTATTGAATTCTGGTCCAATGTGAGCGATGAGGAAATAGATCTCGCAATTGAAGAAGCTGAAGCAGTTGAAGCAGGTCGACCTCCAACTAGAACTTCACGATTCTATGCAAGAGGGGCACTTCAATACCTTGCACCTGTCTTGATGCAAAAACTAACAAAACAAGATGATTCTGATGATGAATTGGAATGGAATCCGTCAAAGGCTGCTTCTGTTTGTCTTATGCTTTTGTCAAATTGCTGTGAAGACGAAATTGTACCGCATGTGTTACCGTTTATTAATTGTAACATTAAAAGTGAAAATTGGCGATACAGAGAAGCTGCTCTAATGGCATTTGGTTCAATTCTTGGTGGTCTTGGAGCTAACACTCTGAAACCATTAGTGGAAAATGCCATGCCAACTTTGATTGAAGCGATGTATGATTCTAGTATAGCTGTAAGGGACACCGCTGCTTGGACATTTGGTAGAATTTGTGAAATTGTCCCTGAAGCTGCTATCAATGAAACATATTTGAAACCATTATTAGAGAGTCTTGTAAATGGTTTAAAAGCTGAACCTAGAGTTGCAGCTAATGTTTGTTGGGCATTCACCGGTTTGGCAGAAGCAGCATACGAGGCAGCGGATGGGGGTGATTCTAATCAGCCAAAGACATATTGCATGTCAAATTATTTTGATTTCATCGTTCAGAGATTACTTGAAACAACGGATCGTCAAGATGCTGCTCAACATAATTTGAGATCAGCTGCATATGAAGCTCTAATGGAAATGGTCAAAAATTCGCCCAGTGACTGTTATGTCACAGTCCAAAAGACCACTATGGTTATACTTGAGAGACTCCAACAAGTTCTACAAATGGAAATACATGTTTCTAGTCAAGCTGACCGTTCTCAATTTAACGACCTCCAAAGTTTATTATGTGCTACCTTGCATTCGGTTCTAAAAAAAGTTACTCCTGATGATGCTCCTCAAATATCAGACACAATAATGACTGCTTTGTTAACTGTATTTGCAAGTAATGCTGGAAAAGCAGGCAGTGTTCAGGAGGATGCTCTGATGGCTGTTTCTACTCTTGTTGAAGTCTTAGGAGAAGAGTTTTTAAAATACATGGATACATTCAAAAAATACCTTTATACTGGTCTTAAAAACCACCAGGAGTACCAAGTTTGCATTGCAGCTGTAGGATTGACTGGTGATATCTGTCGCGCCTTGAAAAATAAGATGGCACCCTATTGTGATGAGATAATAGTTTTGTTATTGGAGAATTTAGGTGATCCATCAATTCACCGATCAGTGAAGCCTGAAATATTATCTGTATTTGGAGACATTGCTTTGAGTATTGGTAGCGATTTTGGAAAATATTTTGATGTCGTCATGCAAATGCTATTGCAGGCTAGTAATGCTCAAGTGGACCATAACGACTATGATATGGTTGAATATTTGGGTGAACTCCGCGAAAGCGTTCTGGAAGCATACACTGGAATCATTCAAGGCCTTAAAGGTGTAGGTTCTCAGGTACGACCGGACATCGCTATCGTCGAGCCGCACGTGCCGGCGATCGTTAACTTCATGATTCAGGTTGCGAACGAAGAGGACCGCACCGACGCCCAGATGGCGGTCATATCGGGGCTCATGGGCGACTTGACTAACGCGTTCGGGGCGCACGTGCTGCCCATGCTCGACACCAAGCCGCTCACCGACATGCTGCAGAGATGCGTCGCGTCAAGCAGACCCTCCCTCTTGCCAGCTGATTTGGAATTACCGACCACAGCATTGAAACGCAGCTTGAAACAACATACTGCTGGTAAAATCAATTTTTGA
Protein
MHAETTLTLIQILEKTVSPDRNELEAAVRYLDHAATTNFTTFIKMLSDVLLQGGNSQVARMAAGLQLKNHLTSKDPTLKQQYQQRWLALAEDVRLYIKENILAAIGTENSRPSSAAQCVAYVAVAELPVGQWNDLIPILVENVVHVQSTELKKEATLEAIGYICQDIDAEVLTERSNQILTAIIHGMRSTEPSNHVRLAATQALLNSLEFTRANFEKENERNFIMEVVCEATQSSDMRISVAALQCLVKILSLYYQHMEPYMGQALFPITLEAMKSDIDEISLQGIEFWSNVSDEEIDLAIEEAEAVEAGRPPTRTSRFYARGALQYLAPVLMQKLTKQDDSDDELEWNPSKAASVCLMLLSNCCEDEIVPHVLPFINCNIKSENWRYREAALMAFGSILGGLGANTLKPLVENAMPTLIEAMYDSSIAVRDTAAWTFGRICEIVPEAAINETYLKPLLESLVNGLKAEPRVAANVCWAFTGLAEAAYEAADGGDSNQPKTYCMSNYFDFIVQRLLETTDRQDAAQHNLRSAAYEALMEMVKNSPSDCYVTVQKTTMVILERLQQVLQMEIHVSSQADRSQFNDLQSLLCATLHSVLKKVTPDDAPQISDTIMTALLTVFASNAGKAGSVQEDALMAVSTLVEVLGEEFLKYMDTFKKYLYTGLKNHQEYQVCIAAVGLTGDICRALKNKMAPYCDEIIVLLLENLGDPSIHRSVKPEILSVFGDIALSIGSDFGKYFDVVMQMLLQASNAQVDHNDYDMVEYLGELRESVLEAYTGIIQGLKGVGSQVRPDIAIVEPHVPAIVNFMIQVANEEDRTDAQMAVISGLMGDLTNAFGAHVLPMLDTKPLTDMLQRCVASSRPSLLPADLELPTTALKRSLKQHTAGKINF
Summary
Uniprot
H9J0D6
A0A2A4K819
A0A2H1VCV7
A0A194QLU9
A0A1E1WEE3
S4PBE6
+ More
A0A212FFM1 A0A2J7RFX1 A0A2J7RFW8 A0A1B6I258 A0A1B6I865 A0A1Y1KG22 D6WNI5 A0A1W4XAG9 E9IJT9 A0A0J7KMD2 E2BR28 A0A1B6BYM1 F4X0K9 K7J9Z2 A0A158ND11 Q0IE87 Q0IE88 A0A1Q3EYW7 A0A1Q3EYE3 B0WBR4 A0A1S4JBY5 A0A3L8DKZ0 A0A0C9RWX3 U5EL75 A0A336LK41 U4UTN2 N6U3L0 A0A087ZMX9 A0A2A3E9L6 A0A195B8V4 A0A151WYU3 A0A0A9WWE9 A0A0M8ZQU5 A0A195D0J7 A0A0K8TFT6 A0A0A9X169 A0A0K8TH25 A0A195D8P1 A0A154P5L2 A0A026WJI9 E2A048 A0A195F7M8 A0A1B0GIQ2 A0A1S4H2E8 A0A3F2YYB8 A0A182KYW9 A0A182XLT9 A0A182VBX3 A0A182TV19 Q7Q186 A0A182HRG5 A0A1L8DXL0 A0A182K1F4 A0A182RR93 A0A182YMR1 A0A069DXP2 A0A182Q9C0 A0A182NAT8 A0A182MDF9 A0A182IPL8 A0A182W8Z5 A0A2M4BDG4 B4JAJ7 A0A0A1XB77 W8C7E3 A0A2M3Z154 T1IDG3 A0A0V0G8U5 A0A023F414 A0A2M4A3T7 W5JHY1 A0A1I8N3Z5 A0A084W8P7 A0A1I8P7H3 A0A182F1N9 A0A224X6X6 A0A034UZ33 A0A182PG28 T1J2G6 A0A0L0C9D4 A0A1S3IUZ5 A0A0M3QUB0 E0VS40 B4MWC0 B4LV89 B4KJ03 B3MK19 A0A1B0D267 V5HPD2 A0A131XZE1 A0A3B0JVU8 A0A1W4UVJ2 E9GU06 A0A1A9XKX3
A0A212FFM1 A0A2J7RFX1 A0A2J7RFW8 A0A1B6I258 A0A1B6I865 A0A1Y1KG22 D6WNI5 A0A1W4XAG9 E9IJT9 A0A0J7KMD2 E2BR28 A0A1B6BYM1 F4X0K9 K7J9Z2 A0A158ND11 Q0IE87 Q0IE88 A0A1Q3EYW7 A0A1Q3EYE3 B0WBR4 A0A1S4JBY5 A0A3L8DKZ0 A0A0C9RWX3 U5EL75 A0A336LK41 U4UTN2 N6U3L0 A0A087ZMX9 A0A2A3E9L6 A0A195B8V4 A0A151WYU3 A0A0A9WWE9 A0A0M8ZQU5 A0A195D0J7 A0A0K8TFT6 A0A0A9X169 A0A0K8TH25 A0A195D8P1 A0A154P5L2 A0A026WJI9 E2A048 A0A195F7M8 A0A1B0GIQ2 A0A1S4H2E8 A0A3F2YYB8 A0A182KYW9 A0A182XLT9 A0A182VBX3 A0A182TV19 Q7Q186 A0A182HRG5 A0A1L8DXL0 A0A182K1F4 A0A182RR93 A0A182YMR1 A0A069DXP2 A0A182Q9C0 A0A182NAT8 A0A182MDF9 A0A182IPL8 A0A182W8Z5 A0A2M4BDG4 B4JAJ7 A0A0A1XB77 W8C7E3 A0A2M3Z154 T1IDG3 A0A0V0G8U5 A0A023F414 A0A2M4A3T7 W5JHY1 A0A1I8N3Z5 A0A084W8P7 A0A1I8P7H3 A0A182F1N9 A0A224X6X6 A0A034UZ33 A0A182PG28 T1J2G6 A0A0L0C9D4 A0A1S3IUZ5 A0A0M3QUB0 E0VS40 B4MWC0 B4LV89 B4KJ03 B3MK19 A0A1B0D267 V5HPD2 A0A131XZE1 A0A3B0JVU8 A0A1W4UVJ2 E9GU06 A0A1A9XKX3
Pubmed
19121390
26354079
23622113
22118469
28004739
18362917
+ More
19820115 21282665 20798317 21719571 20075255 21347285 17510324 30249741 23537049 25401762 26823975 24508170 12364791 20966253 25244985 26334808 17994087 25830018 24495485 25474469 20920257 23761445 25315136 24438588 25348373 26108605 20566863 18057021 25765539 21292972
19820115 21282665 20798317 21719571 20075255 21347285 17510324 30249741 23537049 25401762 26823975 24508170 12364791 20966253 25244985 26334808 17994087 25830018 24495485 25474469 20920257 23761445 25315136 24438588 25348373 26108605 20566863 18057021 25765539 21292972
EMBL
BABH01010537
NWSH01000081
PCG79802.1
ODYU01001874
SOQ38688.1
KQ461196
+ More
KPJ06502.1 GDQN01005729 JAT85325.1 GAIX01004656 JAA87904.1 AGBW02008803 OWR52529.1 NEVH01004408 PNF39734.1 PNF39735.1 GECU01026712 JAS80994.1 GECU01024609 JAS83097.1 GEZM01084868 GEZM01084867 JAV60452.1 KQ971343 EFA04374.2 GL763883 EFZ19149.1 LBMM01005580 KMQ91404.1 GL449898 EFN81834.1 GEDC01030931 GEDC01018372 JAS06367.1 JAS18926.1 GL888498 EGI60001.1 AAZX01002269 AAZX01002270 AAZX01002272 AAZX01004221 AAZX01006609 AAZX01009899 AAZX01011937 AAZX01012571 AAZX01013111 AAZX01015396 ADTU01012159 CH478001 EAT34489.1 EAT34490.1 GFDL01014555 JAV20490.1 GFDL01014726 JAV20319.1 DS231881 EDS42685.1 QOIP01000007 RLU20579.1 GBYB01013570 JAG83337.1 GANO01001534 JAB58337.1 UFQT01000026 SSX18055.1 KB632357 ERL93500.1 APGK01043832 KB741018 ENN75206.1 KZ288335 PBC27866.1 KQ976558 KYM80670.1 KQ982650 KYQ52865.1 GBHO01032757 GBHO01032756 GDHC01011785 JAG10847.1 JAG10848.1 JAQ06844.1 KQ435896 KOX69255.1 KQ977012 KYN06397.1 GBRD01001368 JAG64453.1 GBHO01032754 JAG10850.1 GBRD01001366 JAG64455.1 KQ981135 KYN09243.1 KQ434822 KZC07143.1 KK107213 EZA55289.1 GL435551 EFN73181.1 KQ981744 KYN36386.1 AJWK01015250 AJWK01015251 AJWK01015252 AJWK01015253 AAAB01008980 EAA14059.3 APCN01001701 GFDF01002952 JAV11132.1 GBGD01000417 JAC88472.1 AXCN02000659 AXCM01000135 GGFJ01001860 MBW51001.1 CH916368 EDW02783.1 GBXI01006036 JAD08256.1 GAMC01003645 GAMC01003644 JAC02912.1 GGFM01001499 MBW22250.1 ACPB03012769 GECL01001839 JAP04285.1 GBBI01002998 JAC15714.1 GGFK01002094 MBW35415.1 ADMH02001134 ETN63982.1 ATLV01021497 KE525319 KFB46591.1 GFTR01008219 JAW08207.1 GAKP01023414 JAC35544.1 JH431806 JRES01000714 KNC29033.1 CP012523 ALC40319.1 DS235742 EEB16196.1 CH963857 EDW75990.1 CH940649 EDW64349.1 KRF81570.1 KRF81571.1 CH933807 EDW13516.1 KRG03798.1 KRG03799.1 CH902620 EDV31437.1 KPU73381.1 KPU73382.1 AJVK01010495 AJVK01010496 GANP01006931 JAB77537.1 GEFM01003182 JAP72614.1 OUUW01000004 SPP79620.1 GL732565 EFX76950.1
KPJ06502.1 GDQN01005729 JAT85325.1 GAIX01004656 JAA87904.1 AGBW02008803 OWR52529.1 NEVH01004408 PNF39734.1 PNF39735.1 GECU01026712 JAS80994.1 GECU01024609 JAS83097.1 GEZM01084868 GEZM01084867 JAV60452.1 KQ971343 EFA04374.2 GL763883 EFZ19149.1 LBMM01005580 KMQ91404.1 GL449898 EFN81834.1 GEDC01030931 GEDC01018372 JAS06367.1 JAS18926.1 GL888498 EGI60001.1 AAZX01002269 AAZX01002270 AAZX01002272 AAZX01004221 AAZX01006609 AAZX01009899 AAZX01011937 AAZX01012571 AAZX01013111 AAZX01015396 ADTU01012159 CH478001 EAT34489.1 EAT34490.1 GFDL01014555 JAV20490.1 GFDL01014726 JAV20319.1 DS231881 EDS42685.1 QOIP01000007 RLU20579.1 GBYB01013570 JAG83337.1 GANO01001534 JAB58337.1 UFQT01000026 SSX18055.1 KB632357 ERL93500.1 APGK01043832 KB741018 ENN75206.1 KZ288335 PBC27866.1 KQ976558 KYM80670.1 KQ982650 KYQ52865.1 GBHO01032757 GBHO01032756 GDHC01011785 JAG10847.1 JAG10848.1 JAQ06844.1 KQ435896 KOX69255.1 KQ977012 KYN06397.1 GBRD01001368 JAG64453.1 GBHO01032754 JAG10850.1 GBRD01001366 JAG64455.1 KQ981135 KYN09243.1 KQ434822 KZC07143.1 KK107213 EZA55289.1 GL435551 EFN73181.1 KQ981744 KYN36386.1 AJWK01015250 AJWK01015251 AJWK01015252 AJWK01015253 AAAB01008980 EAA14059.3 APCN01001701 GFDF01002952 JAV11132.1 GBGD01000417 JAC88472.1 AXCN02000659 AXCM01000135 GGFJ01001860 MBW51001.1 CH916368 EDW02783.1 GBXI01006036 JAD08256.1 GAMC01003645 GAMC01003644 JAC02912.1 GGFM01001499 MBW22250.1 ACPB03012769 GECL01001839 JAP04285.1 GBBI01002998 JAC15714.1 GGFK01002094 MBW35415.1 ADMH02001134 ETN63982.1 ATLV01021497 KE525319 KFB46591.1 GFTR01008219 JAW08207.1 GAKP01023414 JAC35544.1 JH431806 JRES01000714 KNC29033.1 CP012523 ALC40319.1 DS235742 EEB16196.1 CH963857 EDW75990.1 CH940649 EDW64349.1 KRF81570.1 KRF81571.1 CH933807 EDW13516.1 KRG03798.1 KRG03799.1 CH902620 EDV31437.1 KPU73381.1 KPU73382.1 AJVK01010495 AJVK01010496 GANP01006931 JAB77537.1 GEFM01003182 JAP72614.1 OUUW01000004 SPP79620.1 GL732565 EFX76950.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000235965
UP000007266
+ More
UP000192223 UP000036403 UP000008237 UP000007755 UP000002358 UP000005205 UP000008820 UP000002320 UP000279307 UP000030742 UP000019118 UP000005203 UP000242457 UP000078540 UP000075809 UP000053105 UP000078542 UP000078492 UP000076502 UP000053097 UP000000311 UP000078541 UP000092461 UP000075882 UP000076407 UP000075903 UP000075902 UP000007062 UP000075840 UP000075881 UP000075900 UP000076408 UP000075886 UP000075884 UP000075883 UP000075880 UP000075920 UP000001070 UP000015103 UP000000673 UP000095301 UP000030765 UP000095300 UP000069272 UP000075885 UP000037069 UP000085678 UP000092553 UP000009046 UP000007798 UP000008792 UP000009192 UP000007801 UP000092462 UP000268350 UP000192221 UP000000305 UP000092443
UP000192223 UP000036403 UP000008237 UP000007755 UP000002358 UP000005205 UP000008820 UP000002320 UP000279307 UP000030742 UP000019118 UP000005203 UP000242457 UP000078540 UP000075809 UP000053105 UP000078542 UP000078492 UP000076502 UP000053097 UP000000311 UP000078541 UP000092461 UP000075882 UP000076407 UP000075903 UP000075902 UP000007062 UP000075840 UP000075881 UP000075900 UP000076408 UP000075886 UP000075884 UP000075883 UP000075880 UP000075920 UP000001070 UP000015103 UP000000673 UP000095301 UP000030765 UP000095300 UP000069272 UP000075885 UP000037069 UP000085678 UP000092553 UP000009046 UP000007798 UP000008792 UP000009192 UP000007801 UP000092462 UP000268350 UP000192221 UP000000305 UP000092443
Interpro
Gene 3D
ProteinModelPortal
H9J0D6
A0A2A4K819
A0A2H1VCV7
A0A194QLU9
A0A1E1WEE3
S4PBE6
+ More
A0A212FFM1 A0A2J7RFX1 A0A2J7RFW8 A0A1B6I258 A0A1B6I865 A0A1Y1KG22 D6WNI5 A0A1W4XAG9 E9IJT9 A0A0J7KMD2 E2BR28 A0A1B6BYM1 F4X0K9 K7J9Z2 A0A158ND11 Q0IE87 Q0IE88 A0A1Q3EYW7 A0A1Q3EYE3 B0WBR4 A0A1S4JBY5 A0A3L8DKZ0 A0A0C9RWX3 U5EL75 A0A336LK41 U4UTN2 N6U3L0 A0A087ZMX9 A0A2A3E9L6 A0A195B8V4 A0A151WYU3 A0A0A9WWE9 A0A0M8ZQU5 A0A195D0J7 A0A0K8TFT6 A0A0A9X169 A0A0K8TH25 A0A195D8P1 A0A154P5L2 A0A026WJI9 E2A048 A0A195F7M8 A0A1B0GIQ2 A0A1S4H2E8 A0A3F2YYB8 A0A182KYW9 A0A182XLT9 A0A182VBX3 A0A182TV19 Q7Q186 A0A182HRG5 A0A1L8DXL0 A0A182K1F4 A0A182RR93 A0A182YMR1 A0A069DXP2 A0A182Q9C0 A0A182NAT8 A0A182MDF9 A0A182IPL8 A0A182W8Z5 A0A2M4BDG4 B4JAJ7 A0A0A1XB77 W8C7E3 A0A2M3Z154 T1IDG3 A0A0V0G8U5 A0A023F414 A0A2M4A3T7 W5JHY1 A0A1I8N3Z5 A0A084W8P7 A0A1I8P7H3 A0A182F1N9 A0A224X6X6 A0A034UZ33 A0A182PG28 T1J2G6 A0A0L0C9D4 A0A1S3IUZ5 A0A0M3QUB0 E0VS40 B4MWC0 B4LV89 B4KJ03 B3MK19 A0A1B0D267 V5HPD2 A0A131XZE1 A0A3B0JVU8 A0A1W4UVJ2 E9GU06 A0A1A9XKX3
A0A212FFM1 A0A2J7RFX1 A0A2J7RFW8 A0A1B6I258 A0A1B6I865 A0A1Y1KG22 D6WNI5 A0A1W4XAG9 E9IJT9 A0A0J7KMD2 E2BR28 A0A1B6BYM1 F4X0K9 K7J9Z2 A0A158ND11 Q0IE87 Q0IE88 A0A1Q3EYW7 A0A1Q3EYE3 B0WBR4 A0A1S4JBY5 A0A3L8DKZ0 A0A0C9RWX3 U5EL75 A0A336LK41 U4UTN2 N6U3L0 A0A087ZMX9 A0A2A3E9L6 A0A195B8V4 A0A151WYU3 A0A0A9WWE9 A0A0M8ZQU5 A0A195D0J7 A0A0K8TFT6 A0A0A9X169 A0A0K8TH25 A0A195D8P1 A0A154P5L2 A0A026WJI9 E2A048 A0A195F7M8 A0A1B0GIQ2 A0A1S4H2E8 A0A3F2YYB8 A0A182KYW9 A0A182XLT9 A0A182VBX3 A0A182TV19 Q7Q186 A0A182HRG5 A0A1L8DXL0 A0A182K1F4 A0A182RR93 A0A182YMR1 A0A069DXP2 A0A182Q9C0 A0A182NAT8 A0A182MDF9 A0A182IPL8 A0A182W8Z5 A0A2M4BDG4 B4JAJ7 A0A0A1XB77 W8C7E3 A0A2M3Z154 T1IDG3 A0A0V0G8U5 A0A023F414 A0A2M4A3T7 W5JHY1 A0A1I8N3Z5 A0A084W8P7 A0A1I8P7H3 A0A182F1N9 A0A224X6X6 A0A034UZ33 A0A182PG28 T1J2G6 A0A0L0C9D4 A0A1S3IUZ5 A0A0M3QUB0 E0VS40 B4MWC0 B4LV89 B4KJ03 B3MK19 A0A1B0D267 V5HPD2 A0A131XZE1 A0A3B0JVU8 A0A1W4UVJ2 E9GU06 A0A1A9XKX3
PDB
1UKL
E-value=0,
Score=2761
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
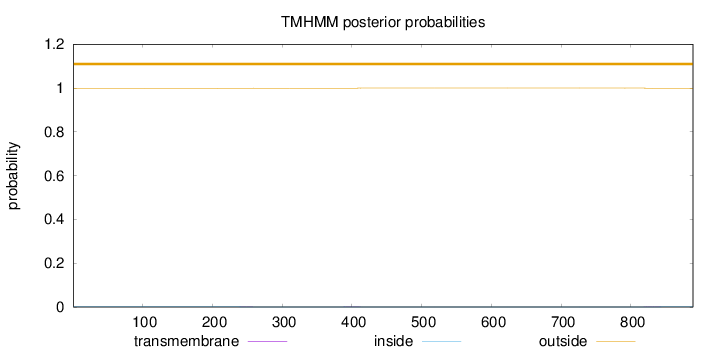
Length:
889
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04258
Exp number, first 60 AAs:
0.00185
Total prob of N-in:
0.00118
outside
1 - 889
Population Genetic Test Statistics
Pi
3.990137
Theta
162.451919
Tajima's D
-1.289472
CLR
2418.126325
CSRT
0.0885455727213639
Interpretation
Uncertain
