Gene
KWMTBOMO02056
Pre Gene Modal
BGIBMGA002970
Annotation
PREDICTED:_uncharacterized_protein_LOC106720006_isoform_X2_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.306 Nuclear Reliability : 1.331 PlasmaMembrane Reliability : 1.08
Sequence
CDS
ATGTGGTATCAAGAGGTAGAAGACGAAGATGAGGACCTTATGTGTTCGCCGGCGATCCTCGCCCGACGTGCCTCCGAAAGCTGGATAAATGTACCTCCAGTTGAGATGCTGAGCACGCCGCAGACGCTTCAACGCAAGAAGTCTCTCCCCGATGTAGCGAGTATGCCGCGGGCGGCGGAGACCGGTATGTCCCGCGAAGAGGTATCGGCGCTAGGATCCGCGCGACGCGAGGAAGTGCGCCGCATGCACGAGGAATCGGAAAAACTGAGGGCCAATCCTCTACTCTACTTGGTCAGTCCGCAAGTCAAGGACTGGTTCTCCCGACAGCAACTGGTTATCCTTGTACTGTTTATCAATATCTCTCTGGGCATCATGTTCTTCAAGCTGCTCACGTAG
Protein
MWYQEVEDEDEDLMCSPAILARRASESWINVPPVEMLSTPQTLQRKKSLPDVASMPRAAETGMSREEVSALGSARREEVRRMHEESEKLRANPLLYLVSPQVKDWFSRQQLVILVLFINISLGIMFFKLLT
Summary
Uniprot
A0A194QSI7
I4DK35
A0A194PJY3
A0A1Y1KJR4
A0A1B6GJC8
A0A1Q3FZ64
+ More
A0A182GZ98 A0A182VIQ5 W5JSU8 T1HGT7 A0A139WHY6 A0A1B6LRJ6 A0A1I8NSD2 A0A1I8MPJ8 D3TS65 A0A1L8D9R3 E2AF02 A0A1B6CE10 A0A0A1XC71 A0A182FNT8 A0A146LIP2 A0A088A1N6 F4WLZ8 A0A182NMX6 A0A0A9WAQ6 A0A026WCG9 A0A0Q9XC78 A0A182HZQ4 A0A1B0FDF1 A0A0R3NSC3 A0A0P8XXT6 A0A1W4VHU5 B4Q7T6 A0A0R1DKU0 B4HYY8 A0A0Q5WAC5 Q6AWN6 A0A0C9QW55 A0A0Q9X142 A0A0Q9W9T7 A0A034WPL0 A0A2A4K706 A0A2A4K6E6 A0A1J1HTH5 A0A182XGM2 A0A2H1V171 A0A182VYF7 A0A0K8W9Q7 Q5TX81 E0VJM2 A0A1W4XQH0 A0A158NEI1 A0A1Y1KES4 A0A195BD23 N6TIB5 U4UJX7 A0A1B6I4S1 A0A1B6J4G8 A0A0M4EE38 A0A1B6F4F4 A0A1B6L2U2 A0A2S2PMF1 A0A139WHB8 A0A0L7LPR6 A0A023F4X2 A0A2S2QRG3 A0A1I8MNI8 A0A0A1XI78 A0A1Z5LCC3 A0A182TIF0 A0A154PP73 A0A0P6HK18 A0A2A3EH95 E9FUC2 A0A3L8DQZ8 A0A151X9W2
A0A182GZ98 A0A182VIQ5 W5JSU8 T1HGT7 A0A139WHY6 A0A1B6LRJ6 A0A1I8NSD2 A0A1I8MPJ8 D3TS65 A0A1L8D9R3 E2AF02 A0A1B6CE10 A0A0A1XC71 A0A182FNT8 A0A146LIP2 A0A088A1N6 F4WLZ8 A0A182NMX6 A0A0A9WAQ6 A0A026WCG9 A0A0Q9XC78 A0A182HZQ4 A0A1B0FDF1 A0A0R3NSC3 A0A0P8XXT6 A0A1W4VHU5 B4Q7T6 A0A0R1DKU0 B4HYY8 A0A0Q5WAC5 Q6AWN6 A0A0C9QW55 A0A0Q9X142 A0A0Q9W9T7 A0A034WPL0 A0A2A4K706 A0A2A4K6E6 A0A1J1HTH5 A0A182XGM2 A0A2H1V171 A0A182VYF7 A0A0K8W9Q7 Q5TX81 E0VJM2 A0A1W4XQH0 A0A158NEI1 A0A1Y1KES4 A0A195BD23 N6TIB5 U4UJX7 A0A1B6I4S1 A0A1B6J4G8 A0A0M4EE38 A0A1B6F4F4 A0A1B6L2U2 A0A2S2PMF1 A0A139WHB8 A0A0L7LPR6 A0A023F4X2 A0A2S2QRG3 A0A1I8MNI8 A0A0A1XI78 A0A1Z5LCC3 A0A182TIF0 A0A154PP73 A0A0P6HK18 A0A2A3EH95 E9FUC2 A0A3L8DQZ8 A0A151X9W2
Pubmed
26354079
22651552
28004739
26483478
20920257
23761445
+ More
18362917 19820115 25315136 20353571 20798317 25830018 26823975 21719571 25401762 24508170 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 12364791 14747013 17210077 20566863 21347285 23537049 26227816 25474469 28528879 21292972 30249741
18362917 19820115 25315136 20353571 20798317 25830018 26823975 21719571 25401762 24508170 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 12364791 14747013 17210077 20566863 21347285 23537049 26227816 25474469 28528879 21292972 30249741
EMBL
KQ461196
KPJ06506.1
AK401653
BAM18275.1
KQ459601
KPI93746.1
+ More
GEZM01086718 JAV59017.1 GECZ01009487 GECZ01007259 JAS60282.1 JAS62510.1 GFDL01002186 JAV32859.1 JXUM01099089 JXUM01099090 JXUM01099091 JXUM01099092 KQ564468 KXJ72230.1 ADMH02000189 ETN67432.1 ACPB03028380 KQ971343 KYB27387.1 GEBQ01013798 JAT26179.1 EZ424267 ADD20543.1 GFDF01010868 JAV03216.1 GL438984 EFN67957.1 GEDC01025612 JAS11686.1 GBXI01005750 JAD08542.1 GDHC01010678 JAQ07951.1 GL888217 EGI64642.1 GBHO01038725 GBRD01016814 JAG04879.1 JAG49013.1 KK107293 EZA53376.1 CH933807 KRG03162.1 APCN01001998 CCAG010016514 CH379060 KRT03836.1 CH902624 KPU74258.1 CM000361 EDX04368.1 CM000157 KRJ97963.1 CH480818 EDW52268.1 CH954177 KQS70380.1 BT015212 AE014134 KX531630 AAT94441.1 ABV53660.1 ANY27440.1 GBYB01007969 JAG77736.1 CH963857 KRF98498.1 CH940654 KRF77972.1 GAKP01002824 GAKP01002822 JAC56130.1 NWSH01000081 PCG79806.1 PCG79807.1 CVRI01000020 CRK91301.1 ODYU01000205 SOQ34595.1 GDHF01032918 GDHF01004423 JAI19396.1 JAI47891.1 AAAB01008799 EAL42041.4 DS235226 EEB13578.1 ADTU01001390 ADTU01001391 GEZM01086716 JAV59018.1 KQ976519 KYM82112.1 APGK01025469 APGK01025470 KB740591 ENN80159.1 KB632226 ERL90300.1 GECU01025802 JAS81904.1 GECU01013634 JAS94072.1 CP012523 ALC40222.1 GECZ01027710 GECZ01024714 JAS42059.1 JAS45055.1 GEBQ01022011 JAT17966.1 GGMR01018003 MBY30622.1 KYB27388.1 JTDY01000385 KOB77435.1 GBBI01002634 JAC16078.1 GGMS01011163 MBY80366.1 GBXI01003218 JAD11074.1 GFJQ02001907 JAW05063.1 KQ434978 KZC12970.1 GDIQ01018008 JAN76729.1 KZ288250 PBC31097.1 GL732525 EFX88946.1 QOIP01000005 RLU22309.1 KQ982353 KYQ57162.1
GEZM01086718 JAV59017.1 GECZ01009487 GECZ01007259 JAS60282.1 JAS62510.1 GFDL01002186 JAV32859.1 JXUM01099089 JXUM01099090 JXUM01099091 JXUM01099092 KQ564468 KXJ72230.1 ADMH02000189 ETN67432.1 ACPB03028380 KQ971343 KYB27387.1 GEBQ01013798 JAT26179.1 EZ424267 ADD20543.1 GFDF01010868 JAV03216.1 GL438984 EFN67957.1 GEDC01025612 JAS11686.1 GBXI01005750 JAD08542.1 GDHC01010678 JAQ07951.1 GL888217 EGI64642.1 GBHO01038725 GBRD01016814 JAG04879.1 JAG49013.1 KK107293 EZA53376.1 CH933807 KRG03162.1 APCN01001998 CCAG010016514 CH379060 KRT03836.1 CH902624 KPU74258.1 CM000361 EDX04368.1 CM000157 KRJ97963.1 CH480818 EDW52268.1 CH954177 KQS70380.1 BT015212 AE014134 KX531630 AAT94441.1 ABV53660.1 ANY27440.1 GBYB01007969 JAG77736.1 CH963857 KRF98498.1 CH940654 KRF77972.1 GAKP01002824 GAKP01002822 JAC56130.1 NWSH01000081 PCG79806.1 PCG79807.1 CVRI01000020 CRK91301.1 ODYU01000205 SOQ34595.1 GDHF01032918 GDHF01004423 JAI19396.1 JAI47891.1 AAAB01008799 EAL42041.4 DS235226 EEB13578.1 ADTU01001390 ADTU01001391 GEZM01086716 JAV59018.1 KQ976519 KYM82112.1 APGK01025469 APGK01025470 KB740591 ENN80159.1 KB632226 ERL90300.1 GECU01025802 JAS81904.1 GECU01013634 JAS94072.1 CP012523 ALC40222.1 GECZ01027710 GECZ01024714 JAS42059.1 JAS45055.1 GEBQ01022011 JAT17966.1 GGMR01018003 MBY30622.1 KYB27388.1 JTDY01000385 KOB77435.1 GBBI01002634 JAC16078.1 GGMS01011163 MBY80366.1 GBXI01003218 JAD11074.1 GFJQ02001907 JAW05063.1 KQ434978 KZC12970.1 GDIQ01018008 JAN76729.1 KZ288250 PBC31097.1 GL732525 EFX88946.1 QOIP01000005 RLU22309.1 KQ982353 KYQ57162.1
Proteomes
UP000053240
UP000053268
UP000069940
UP000249989
UP000075903
UP000000673
+ More
UP000015103 UP000007266 UP000095300 UP000095301 UP000000311 UP000069272 UP000005203 UP000007755 UP000075884 UP000053097 UP000009192 UP000075840 UP000092444 UP000001819 UP000007801 UP000192221 UP000000304 UP000002282 UP000001292 UP000008711 UP000000803 UP000007798 UP000008792 UP000218220 UP000183832 UP000076407 UP000075920 UP000007062 UP000009046 UP000192223 UP000005205 UP000078540 UP000019118 UP000030742 UP000092553 UP000037510 UP000075902 UP000076502 UP000242457 UP000000305 UP000279307 UP000075809
UP000015103 UP000007266 UP000095300 UP000095301 UP000000311 UP000069272 UP000005203 UP000007755 UP000075884 UP000053097 UP000009192 UP000075840 UP000092444 UP000001819 UP000007801 UP000192221 UP000000304 UP000002282 UP000001292 UP000008711 UP000000803 UP000007798 UP000008792 UP000218220 UP000183832 UP000076407 UP000075920 UP000007062 UP000009046 UP000192223 UP000005205 UP000078540 UP000019118 UP000030742 UP000092553 UP000037510 UP000075902 UP000076502 UP000242457 UP000000305 UP000279307 UP000075809
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194QSI7
I4DK35
A0A194PJY3
A0A1Y1KJR4
A0A1B6GJC8
A0A1Q3FZ64
+ More
A0A182GZ98 A0A182VIQ5 W5JSU8 T1HGT7 A0A139WHY6 A0A1B6LRJ6 A0A1I8NSD2 A0A1I8MPJ8 D3TS65 A0A1L8D9R3 E2AF02 A0A1B6CE10 A0A0A1XC71 A0A182FNT8 A0A146LIP2 A0A088A1N6 F4WLZ8 A0A182NMX6 A0A0A9WAQ6 A0A026WCG9 A0A0Q9XC78 A0A182HZQ4 A0A1B0FDF1 A0A0R3NSC3 A0A0P8XXT6 A0A1W4VHU5 B4Q7T6 A0A0R1DKU0 B4HYY8 A0A0Q5WAC5 Q6AWN6 A0A0C9QW55 A0A0Q9X142 A0A0Q9W9T7 A0A034WPL0 A0A2A4K706 A0A2A4K6E6 A0A1J1HTH5 A0A182XGM2 A0A2H1V171 A0A182VYF7 A0A0K8W9Q7 Q5TX81 E0VJM2 A0A1W4XQH0 A0A158NEI1 A0A1Y1KES4 A0A195BD23 N6TIB5 U4UJX7 A0A1B6I4S1 A0A1B6J4G8 A0A0M4EE38 A0A1B6F4F4 A0A1B6L2U2 A0A2S2PMF1 A0A139WHB8 A0A0L7LPR6 A0A023F4X2 A0A2S2QRG3 A0A1I8MNI8 A0A0A1XI78 A0A1Z5LCC3 A0A182TIF0 A0A154PP73 A0A0P6HK18 A0A2A3EH95 E9FUC2 A0A3L8DQZ8 A0A151X9W2
A0A182GZ98 A0A182VIQ5 W5JSU8 T1HGT7 A0A139WHY6 A0A1B6LRJ6 A0A1I8NSD2 A0A1I8MPJ8 D3TS65 A0A1L8D9R3 E2AF02 A0A1B6CE10 A0A0A1XC71 A0A182FNT8 A0A146LIP2 A0A088A1N6 F4WLZ8 A0A182NMX6 A0A0A9WAQ6 A0A026WCG9 A0A0Q9XC78 A0A182HZQ4 A0A1B0FDF1 A0A0R3NSC3 A0A0P8XXT6 A0A1W4VHU5 B4Q7T6 A0A0R1DKU0 B4HYY8 A0A0Q5WAC5 Q6AWN6 A0A0C9QW55 A0A0Q9X142 A0A0Q9W9T7 A0A034WPL0 A0A2A4K706 A0A2A4K6E6 A0A1J1HTH5 A0A182XGM2 A0A2H1V171 A0A182VYF7 A0A0K8W9Q7 Q5TX81 E0VJM2 A0A1W4XQH0 A0A158NEI1 A0A1Y1KES4 A0A195BD23 N6TIB5 U4UJX7 A0A1B6I4S1 A0A1B6J4G8 A0A0M4EE38 A0A1B6F4F4 A0A1B6L2U2 A0A2S2PMF1 A0A139WHB8 A0A0L7LPR6 A0A023F4X2 A0A2S2QRG3 A0A1I8MNI8 A0A0A1XI78 A0A1Z5LCC3 A0A182TIF0 A0A154PP73 A0A0P6HK18 A0A2A3EH95 E9FUC2 A0A3L8DQZ8 A0A151X9W2
Ontologies
GO
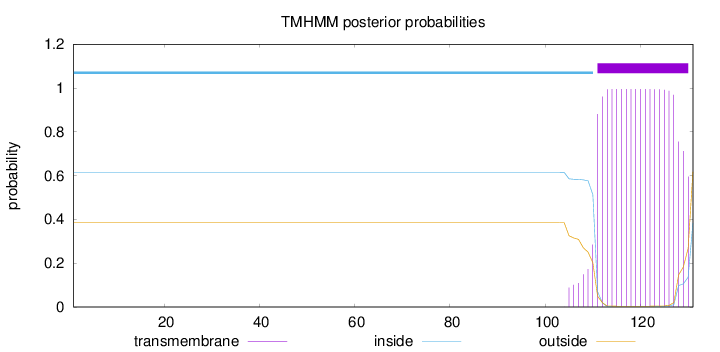
Topology
Length:
131
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.69535
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.61489
inside
1 - 110
TMhelix
111 - 130
outside
131 - 131
Population Genetic Test Statistics
Pi
214.116786
Theta
163.821746
Tajima's D
0.865004
CLR
0.011448
CSRT
0.625518724063797
Interpretation
Uncertain
