Gene
KWMTBOMO02054
Pre Gene Modal
BGIBMGA003198
Annotation
PREDICTED:_major_facilitator_superfamily_domain-containing_protein_1-like_isoform_X1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.985
Sequence
CDS
ATGGATAATCCCGAGGCTAGTGGGAGTAGTAGAACTCCACAGATATCACCCACATCATGGCGCCATCCTGCAAATAGGATACATCGGTTTATCGCTCTTCTGTTAATGTGCTTTCTTTGTTTTGGTTCATATTTCTGTTACGACACACCAGGTGCCCTCGTAGATAACTTCAAAGATGACACACATTTGACAACTTCCCAGTTTACTTTACTTTATTCTATATATTCATGGCCCAATGTTATATTATGCTTTATTGGTGGATACTTAGTTGACAGATATTTTGGTGTAAGATTAGGAACAGTAATATACATGACCATTGTTTTTATTGGAGCTGTCATATTCGCAAGTGGTGTCTATATAAATACATACTGGCTGATGATACTTGGAAGATTCGTATTTGGTATTGGTGGAGAATCATTACAAGTAGTAGTCAATAATTATGTAGTTCTGTGGTTTAAAGGAAAAGAACTAAACATGGTGTTTGGGCTACAATTATCATTCTCTAGGTTCGGTAGTACTGTGAACTTTTGGATAATGGAACCCATTTATAGATGGATTGGCAACTACTACATAGGCTATGAGCGTTTAGGAGTTACTCTATTTATAGCATCAATGACTTGCTTTGTTTCCCTTCTGTGTGGAATAATATTAGGCTGGATGGACTACAGAGCCGAAAAGATCCTAAACAGACAAGAAGAGAGCATGAACGAGGAACCGTTTCACTTGAAAGACATAATGCACTTCAGATCAGAGTTTTGGCTTATTTCTGTCATAGCTGTTGTATACTATTTAGCTTTATTTCCGTTTATTGCATTAGGAAAATTATTTTTCGAACGGAAATTCGACTTTTCACCTCAAGATGCGAACACGGTCAATTCAATGGTTTATCTGTTATCTGCAGCACTGAGTCCCTTTTTTGGCATCCTGATCGATAAAACTGGTAGAAACGTGACTTGGGTTATTATAAGTATCGTCCTGACCTTGTGTGGACATTTCTTGATGGCATTTACATTCTTCAATCCTTATATAGGTGTCATTTTATTAGGAATTTCTTACTCATTATTGTCGAGTGGTTTGTGGCCCTTGGTCGCGCTGATTGTACCCGAGAGTCAACTGGGAACTGCGTATGGAATATGTCAGGCAGTACAAAATATGGGATTAGCAGTGGTCATCATTGCGGCGGGAGTCATCGTCGATAAATACGGATACTTAATGTTGGAAATGTTCTTCCTCGGCTGTCTTTTTATTTCACTGATAGCTGCTGTCTTCATTTACATAATCGATTCTGCGAACAACGGTATTTTGAATATGTCTCCAAATATGCGAGAAAGTACGAAGAGTCAAAGGAGAGTCGATGGTGTTGGTGAAACAGCAAATTTGTTAGAAAATGAAGACTTTACTGATCAAGAAGAAACTGACTCTCGAGTTCAAGAGGAGAAACAGCCTTCAGTCGCTCCACGTGACTCCGCTCTGGTCCAGTCAGACAGAATACGATCCAGATACTTAGCACAAATATTGCCAAATACTAACATTCCAGAACGACAATAA
Protein
MDNPEASGSSRTPQISPTSWRHPANRIHRFIALLLMCFLCFGSYFCYDTPGALVDNFKDDTHLTTSQFTLLYSIYSWPNVILCFIGGYLVDRYFGVRLGTVIYMTIVFIGAVIFASGVYINTYWLMILGRFVFGIGGESLQVVVNNYVVLWFKGKELNMVFGLQLSFSRFGSTVNFWIMEPIYRWIGNYYIGYERLGVTLFIASMTCFVSLLCGIILGWMDYRAEKILNRQEESMNEEPFHLKDIMHFRSEFWLISVIAVVYYLALFPFIALGKLFFERKFDFSPQDANTVNSMVYLLSAALSPFFGILIDKTGRNVTWVIISIVLTLCGHFLMAFTFFNPYIGVILLGISYSLLSSGLWPLVALIVPESQLGTAYGICQAVQNMGLAVVIIAAGVIVDKYGYLMLEMFFLGCLFISLIAAVFIYIIDSANNGILNMSPNMRESTKSQRRVDGVGETANLLENEDFTDQEETDSRVQEEKQPSVAPRDSALVQSDRIRSRYLAQILPNTNIPERQ
Summary
Similarity
Belongs to the enoyl-CoA hydratase/isomerase family.
Uniprot
H9J111
A0A194PK97
S4P7H0
A0A2H1WAM2
A0A212FFM4
A0A194QMC6
+ More
A0A0L7LQF4 Q177X1 U5ET16 A0A1Q3FER1 A0A1S4J601 A0A182P939 A0A336MK01 A0A182VV03 A0A182RGG4 A0A0M9A109 A0A182Q120 A0A182Y5T3 A0A182VA22 A0A182X7P0 A0A182NED8 A0A182IE29 A0A182L5P3 A0A182K823 A0A1Q3FBM4 A0A1Q3FBZ9 A0A1L8DE70 Q177X0 B0W5R5 A0A182TW58 A0A2A3EI64 A0A182JBJ1 A0A088ATF9 Q7PSA6 A0A1B0CJJ6 A0A182MQY3 A0A084VPD6 A0A2M4A4E9 A0A2M3Z452 A0A182F5W9 W5JJA9 E2B3L8 A0A2M4BJQ7 A0A2M4BJX8 A0A195E5D0 A0A026WRR7 A0A195EUD4 A0A1J1I378 A0A158NG96 A0A151WM38 F4WP54 A0A151IL59 A0A154NX98 E2A3K0 E0VY61 A0A0L7RK84 A0A310S6W7 A0A2J7PHE0 A0A1Y0AWP9 W8ARX2 A0A1W7R5H9 A0A1Y1LQE0 A0A1I8Q429 K7INF1 A0A232EMP6 B3N4A4 W8BQ15 A0A034V7J1 T1PLX6 A0A1B0DB20 A0A0L0CG77 A0A182S8L9 A0A034VBW1 A0A0Q9X8I6 B4KGP9 A0A0J9RQF0 B4HV44 A0A0R1DQU5 A0A1B0APS5 B3NFF6 D3TLU4 D6WLP8 B4NZ16 A0A139WH97 Q9VS47 A0A1A9VXR8 A0A1A9YG79 A0A0M4E3H4 B4J1Z6 B4LC95 B3M565 Q9VR34 B4I3D2 B4Q334 A0A0M3QWM6 A0A1A9WMB9 Q1RKW9 A0A0P5B714 A0A0P5X0M0
A0A0L7LQF4 Q177X1 U5ET16 A0A1Q3FER1 A0A1S4J601 A0A182P939 A0A336MK01 A0A182VV03 A0A182RGG4 A0A0M9A109 A0A182Q120 A0A182Y5T3 A0A182VA22 A0A182X7P0 A0A182NED8 A0A182IE29 A0A182L5P3 A0A182K823 A0A1Q3FBM4 A0A1Q3FBZ9 A0A1L8DE70 Q177X0 B0W5R5 A0A182TW58 A0A2A3EI64 A0A182JBJ1 A0A088ATF9 Q7PSA6 A0A1B0CJJ6 A0A182MQY3 A0A084VPD6 A0A2M4A4E9 A0A2M3Z452 A0A182F5W9 W5JJA9 E2B3L8 A0A2M4BJQ7 A0A2M4BJX8 A0A195E5D0 A0A026WRR7 A0A195EUD4 A0A1J1I378 A0A158NG96 A0A151WM38 F4WP54 A0A151IL59 A0A154NX98 E2A3K0 E0VY61 A0A0L7RK84 A0A310S6W7 A0A2J7PHE0 A0A1Y0AWP9 W8ARX2 A0A1W7R5H9 A0A1Y1LQE0 A0A1I8Q429 K7INF1 A0A232EMP6 B3N4A4 W8BQ15 A0A034V7J1 T1PLX6 A0A1B0DB20 A0A0L0CG77 A0A182S8L9 A0A034VBW1 A0A0Q9X8I6 B4KGP9 A0A0J9RQF0 B4HV44 A0A0R1DQU5 A0A1B0APS5 B3NFF6 D3TLU4 D6WLP8 B4NZ16 A0A139WH97 Q9VS47 A0A1A9VXR8 A0A1A9YG79 A0A0M4E3H4 B4J1Z6 B4LC95 B3M565 Q9VR34 B4I3D2 B4Q334 A0A0M3QWM6 A0A1A9WMB9 Q1RKW9 A0A0P5B714 A0A0P5X0M0
Pubmed
19121390
26354079
23622113
22118469
26227816
17510324
+ More
25244985 20966253 12364791 24438588 20920257 23761445 20798317 24508170 30249741 21347285 21719571 20566863 28341416 24495485 28004739 20075255 28648823 17994087 18057021 25348373 25315136 26108605 22936249 17550304 20353571 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
25244985 20966253 12364791 24438588 20920257 23761445 20798317 24508170 30249741 21347285 21719571 20566863 28341416 24495485 28004739 20075255 28648823 17994087 18057021 25348373 25315136 26108605 22936249 17550304 20353571 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01010544
BABH01010545
KQ459601
KPI93747.1
GAIX01004619
JAA87941.1
+ More
ODYU01007388 SOQ50103.1 AGBW02008803 OWR52532.1 KQ461196 KPJ06509.1 JTDY01000385 KOB77441.1 CH477370 EAT42455.1 GANO01002138 JAB57733.1 GFDL01009017 JAV26028.1 UFQS01001478 UFQT01001478 SSX11162.1 SSX30732.1 KQ435771 KOX75155.1 AXCN02000860 APCN01004556 GFDL01010085 JAV24960.1 GFDL01010027 JAV25018.1 GFDF01009332 JAV04752.1 EAT42454.1 DS231844 EDS35718.1 KZ288253 PBC30962.1 AAAB01008839 EAA05922.5 AJWK01014674 AJWK01014675 AJWK01014676 AJWK01014677 AJWK01014678 AXCM01007640 ATLV01014998 KE524999 KFB39830.1 GGFK01002365 MBW35686.1 GGFM01002522 MBW23273.1 ADMH02001016 ETN64447.1 GL445358 EFN89704.1 GGFJ01004121 MBW53262.1 GGFJ01004120 MBW53261.1 KQ979608 KYN20395.1 KK107139 QOIP01000003 EZA57799.1 RLU24694.1 KQ981965 KYN31766.1 CVRI01000038 CRK94234.1 ADTU01015079 KQ982944 KYQ48934.1 GL888243 EGI63985.1 KQ977164 KYN05221.1 KQ434777 KZC04201.1 GL436444 EFN71982.1 DS235843 EEB18317.1 KQ414578 KOC71151.1 KQ797156 OAD46998.1 NEVH01025138 PNF15757.1 KY921821 ART29410.1 GAMC01014972 GAMC01014970 JAB91585.1 GEHC01001272 JAV46373.1 GEZM01054161 GEZM01054160 GEZM01054159 JAV73816.1 NNAY01003330 OXU19611.1 CH954177 EDV57774.1 KQS70118.1 KQS70119.1 GAMC01014971 GAMC01014969 JAB91584.1 GAKP01019706 GAKP01019704 JAC39246.1 KA649796 AFP64425.1 AJVK01000643 JRES01000438 KNC31255.1 GAKP01019707 GAKP01019705 JAC39245.1 CH933807 KRG03657.1 EDW13249.2 KRG03655.1 KRG03656.1 CM002912 KMY98053.1 CH480817 EDW50815.1 CM000157 KRJ97299.1 JXJN01001569 CH954178 EDV50498.1 EZ422396 ADD18672.1 KQ971343 EFA04148.1 EDW87680.2 KRJ97297.1 KRJ97298.1 KRJ97300.1 KYB27225.1 AE014296 AY069628 AAF50582.1 AAL39773.1 AGB94200.1 CP012523 ALC38171.1 CH916366 EDV95921.1 CH940647 EDW68740.1 KRF83995.1 CH902618 EDV40570.1 AE014134 BT133284 AAF50974.2 AFC62083.1 CH480820 EDW54277.1 CM000361 CM002910 EDX03750.1 KMY88128.1 CP012525 ALC44413.1 BT025091 ABE73262.1 GDIP01188578 JAJ34824.1 GDIP01079436 JAM24279.1
ODYU01007388 SOQ50103.1 AGBW02008803 OWR52532.1 KQ461196 KPJ06509.1 JTDY01000385 KOB77441.1 CH477370 EAT42455.1 GANO01002138 JAB57733.1 GFDL01009017 JAV26028.1 UFQS01001478 UFQT01001478 SSX11162.1 SSX30732.1 KQ435771 KOX75155.1 AXCN02000860 APCN01004556 GFDL01010085 JAV24960.1 GFDL01010027 JAV25018.1 GFDF01009332 JAV04752.1 EAT42454.1 DS231844 EDS35718.1 KZ288253 PBC30962.1 AAAB01008839 EAA05922.5 AJWK01014674 AJWK01014675 AJWK01014676 AJWK01014677 AJWK01014678 AXCM01007640 ATLV01014998 KE524999 KFB39830.1 GGFK01002365 MBW35686.1 GGFM01002522 MBW23273.1 ADMH02001016 ETN64447.1 GL445358 EFN89704.1 GGFJ01004121 MBW53262.1 GGFJ01004120 MBW53261.1 KQ979608 KYN20395.1 KK107139 QOIP01000003 EZA57799.1 RLU24694.1 KQ981965 KYN31766.1 CVRI01000038 CRK94234.1 ADTU01015079 KQ982944 KYQ48934.1 GL888243 EGI63985.1 KQ977164 KYN05221.1 KQ434777 KZC04201.1 GL436444 EFN71982.1 DS235843 EEB18317.1 KQ414578 KOC71151.1 KQ797156 OAD46998.1 NEVH01025138 PNF15757.1 KY921821 ART29410.1 GAMC01014972 GAMC01014970 JAB91585.1 GEHC01001272 JAV46373.1 GEZM01054161 GEZM01054160 GEZM01054159 JAV73816.1 NNAY01003330 OXU19611.1 CH954177 EDV57774.1 KQS70118.1 KQS70119.1 GAMC01014971 GAMC01014969 JAB91584.1 GAKP01019706 GAKP01019704 JAC39246.1 KA649796 AFP64425.1 AJVK01000643 JRES01000438 KNC31255.1 GAKP01019707 GAKP01019705 JAC39245.1 CH933807 KRG03657.1 EDW13249.2 KRG03655.1 KRG03656.1 CM002912 KMY98053.1 CH480817 EDW50815.1 CM000157 KRJ97299.1 JXJN01001569 CH954178 EDV50498.1 EZ422396 ADD18672.1 KQ971343 EFA04148.1 EDW87680.2 KRJ97297.1 KRJ97298.1 KRJ97300.1 KYB27225.1 AE014296 AY069628 AAF50582.1 AAL39773.1 AGB94200.1 CP012523 ALC38171.1 CH916366 EDV95921.1 CH940647 EDW68740.1 KRF83995.1 CH902618 EDV40570.1 AE014134 BT133284 AAF50974.2 AFC62083.1 CH480820 EDW54277.1 CM000361 CM002910 EDX03750.1 KMY88128.1 CP012525 ALC44413.1 BT025091 ABE73262.1 GDIP01188578 JAJ34824.1 GDIP01079436 JAM24279.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000037510
UP000008820
+ More
UP000075885 UP000075920 UP000075900 UP000053105 UP000075886 UP000076408 UP000075903 UP000076407 UP000075884 UP000075840 UP000075882 UP000075881 UP000002320 UP000075902 UP000242457 UP000075880 UP000005203 UP000007062 UP000092461 UP000075883 UP000030765 UP000069272 UP000000673 UP000008237 UP000078492 UP000053097 UP000279307 UP000078541 UP000183832 UP000005205 UP000075809 UP000007755 UP000078542 UP000076502 UP000000311 UP000009046 UP000053825 UP000235965 UP000095300 UP000002358 UP000215335 UP000008711 UP000095301 UP000092462 UP000037069 UP000075901 UP000009192 UP000001292 UP000002282 UP000092460 UP000007266 UP000000803 UP000078200 UP000092443 UP000092553 UP000001070 UP000008792 UP000007801 UP000000304 UP000091820
UP000075885 UP000075920 UP000075900 UP000053105 UP000075886 UP000076408 UP000075903 UP000076407 UP000075884 UP000075840 UP000075882 UP000075881 UP000002320 UP000075902 UP000242457 UP000075880 UP000005203 UP000007062 UP000092461 UP000075883 UP000030765 UP000069272 UP000000673 UP000008237 UP000078492 UP000053097 UP000279307 UP000078541 UP000183832 UP000005205 UP000075809 UP000007755 UP000078542 UP000076502 UP000000311 UP000009046 UP000053825 UP000235965 UP000095300 UP000002358 UP000215335 UP000008711 UP000095301 UP000092462 UP000037069 UP000075901 UP000009192 UP000001292 UP000002282 UP000092460 UP000007266 UP000000803 UP000078200 UP000092443 UP000092553 UP000001070 UP000008792 UP000007801 UP000000304 UP000091820
Interpro
IPR011701
MFS
+ More
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR008927 6-PGluconate_DH-like_C_sf
IPR006108 3HC_DH_C
IPR006176 3-OHacyl-CoA_DH_NAD-bd
IPR029045 ClpP/crotonase-like_dom_sf
IPR036291 NAD(P)-bd_dom_sf
IPR001753 Enoyl-CoA_hydra/iso
IPR012803 Fa_ox_alpha_mit
IPR018376 Enoyl-CoA_hyd/isom_CS
IPR036259 MFS_trans_sf
IPR020846 MFS_dom
IPR008927 6-PGluconate_DH-like_C_sf
IPR006108 3HC_DH_C
IPR006176 3-OHacyl-CoA_DH_NAD-bd
IPR029045 ClpP/crotonase-like_dom_sf
IPR036291 NAD(P)-bd_dom_sf
IPR001753 Enoyl-CoA_hydra/iso
IPR012803 Fa_ox_alpha_mit
IPR018376 Enoyl-CoA_hyd/isom_CS
CDD
ProteinModelPortal
H9J111
A0A194PK97
S4P7H0
A0A2H1WAM2
A0A212FFM4
A0A194QMC6
+ More
A0A0L7LQF4 Q177X1 U5ET16 A0A1Q3FER1 A0A1S4J601 A0A182P939 A0A336MK01 A0A182VV03 A0A182RGG4 A0A0M9A109 A0A182Q120 A0A182Y5T3 A0A182VA22 A0A182X7P0 A0A182NED8 A0A182IE29 A0A182L5P3 A0A182K823 A0A1Q3FBM4 A0A1Q3FBZ9 A0A1L8DE70 Q177X0 B0W5R5 A0A182TW58 A0A2A3EI64 A0A182JBJ1 A0A088ATF9 Q7PSA6 A0A1B0CJJ6 A0A182MQY3 A0A084VPD6 A0A2M4A4E9 A0A2M3Z452 A0A182F5W9 W5JJA9 E2B3L8 A0A2M4BJQ7 A0A2M4BJX8 A0A195E5D0 A0A026WRR7 A0A195EUD4 A0A1J1I378 A0A158NG96 A0A151WM38 F4WP54 A0A151IL59 A0A154NX98 E2A3K0 E0VY61 A0A0L7RK84 A0A310S6W7 A0A2J7PHE0 A0A1Y0AWP9 W8ARX2 A0A1W7R5H9 A0A1Y1LQE0 A0A1I8Q429 K7INF1 A0A232EMP6 B3N4A4 W8BQ15 A0A034V7J1 T1PLX6 A0A1B0DB20 A0A0L0CG77 A0A182S8L9 A0A034VBW1 A0A0Q9X8I6 B4KGP9 A0A0J9RQF0 B4HV44 A0A0R1DQU5 A0A1B0APS5 B3NFF6 D3TLU4 D6WLP8 B4NZ16 A0A139WH97 Q9VS47 A0A1A9VXR8 A0A1A9YG79 A0A0M4E3H4 B4J1Z6 B4LC95 B3M565 Q9VR34 B4I3D2 B4Q334 A0A0M3QWM6 A0A1A9WMB9 Q1RKW9 A0A0P5B714 A0A0P5X0M0
A0A0L7LQF4 Q177X1 U5ET16 A0A1Q3FER1 A0A1S4J601 A0A182P939 A0A336MK01 A0A182VV03 A0A182RGG4 A0A0M9A109 A0A182Q120 A0A182Y5T3 A0A182VA22 A0A182X7P0 A0A182NED8 A0A182IE29 A0A182L5P3 A0A182K823 A0A1Q3FBM4 A0A1Q3FBZ9 A0A1L8DE70 Q177X0 B0W5R5 A0A182TW58 A0A2A3EI64 A0A182JBJ1 A0A088ATF9 Q7PSA6 A0A1B0CJJ6 A0A182MQY3 A0A084VPD6 A0A2M4A4E9 A0A2M3Z452 A0A182F5W9 W5JJA9 E2B3L8 A0A2M4BJQ7 A0A2M4BJX8 A0A195E5D0 A0A026WRR7 A0A195EUD4 A0A1J1I378 A0A158NG96 A0A151WM38 F4WP54 A0A151IL59 A0A154NX98 E2A3K0 E0VY61 A0A0L7RK84 A0A310S6W7 A0A2J7PHE0 A0A1Y0AWP9 W8ARX2 A0A1W7R5H9 A0A1Y1LQE0 A0A1I8Q429 K7INF1 A0A232EMP6 B3N4A4 W8BQ15 A0A034V7J1 T1PLX6 A0A1B0DB20 A0A0L0CG77 A0A182S8L9 A0A034VBW1 A0A0Q9X8I6 B4KGP9 A0A0J9RQF0 B4HV44 A0A0R1DQU5 A0A1B0APS5 B3NFF6 D3TLU4 D6WLP8 B4NZ16 A0A139WH97 Q9VS47 A0A1A9VXR8 A0A1A9YG79 A0A0M4E3H4 B4J1Z6 B4LC95 B3M565 Q9VR34 B4I3D2 B4Q334 A0A0M3QWM6 A0A1A9WMB9 Q1RKW9 A0A0P5B714 A0A0P5X0M0
Ontologies
KEGG
GO
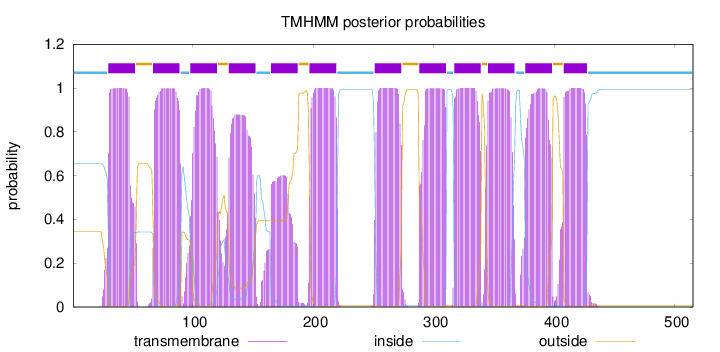
Topology
Length:
515
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
253.95888
Exp number, first 60 AAs:
21.17125
Total prob of N-in:
0.65648
POSSIBLE N-term signal
sequence
inside
1 - 29
TMhelix
30 - 52
outside
53 - 66
TMhelix
67 - 89
inside
90 - 97
TMhelix
98 - 120
outside
121 - 129
TMhelix
130 - 152
inside
153 - 164
TMhelix
165 - 187
outside
188 - 196
TMhelix
197 - 219
inside
220 - 250
TMhelix
251 - 273
outside
274 - 287
TMhelix
288 - 310
inside
311 - 316
TMhelix
317 - 339
outside
340 - 344
TMhelix
345 - 367
inside
368 - 375
TMhelix
376 - 398
outside
399 - 407
TMhelix
408 - 427
inside
428 - 515
Population Genetic Test Statistics
Pi
165.803823
Theta
154.02304
Tajima's D
-0.796684
CLR
0.340069
CSRT
0.173891305434728
Interpretation
Uncertain
