Gene
KWMTBOMO02053 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002967
Annotation
hydroxyacyl-coenzyme_A_dehydrogenase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 3.021
Sequence
CDS
ATGGCGAATAGCAAAATATTAAGTGCCCTTAAGATCTTGAGAAGCCGTAAGGAATTATTCATATCCGGTGTACATAGCCGAAAGTATGCCGTTCCAGCAAGCCAAGTACATACAAAATGCAAATTGGTCAATGGAGTTTACGTAGTCACTTTGGACTCTCCAAATGTTAAAGTAAACTCACTTAACACCCAAGTGATGGAAGAAGTTAGTAACATCGTAAATGAAATTGAAACAAATTCTGGTATTGAAGCTGCAGTCATTATATCAGGCAAGCCTGGATGCTTCATTGCAGGGGCTGATATAAGCATGATCGAAAACTGCAAAACAAAGGAAGAAGTTGTCAGTCTGTCTAAAAGAGGACATGAAATATTCAGAAGAATCGAACAATCACGGAAACCGTATATTGCCGCAATACAAGGCAGCTGTCTAGGAGGTGGATTAGAGACTGCTCTTGCATGTAAATATCGCATTGCTGTGAAAGACTCCAAAACAGGATTTGGACTACCAGAAGTCATGTTGGGACTTTTGCCCGGCGGTGGAGGGACACAGAGATTACCTGCTCTCACCTCCATACCTACAACTTTGGACCTAGCTCTAACTGGTAAAACTGTGAAAGCTGATAAAGCAAAGAAATTGGGAATTGTAGATCTGTTAGTTTCCCCTCTTGGACCAGGTTTGGGTCAGCCTGAGGAAAATACAGCCAAATACCTCGAAACTGTAGCTATACAGATTGCAAAGGATATAGCTGCTGGAAAAATAATTGTTGATAGATCGAAAAAAGGCCTAGTACAGAAGATTACTGCAATCGTAATGCAATGGGAGTTTGTGAAAAACATCATATTTAAGAAAGCCAAGGAACAAGTAATGAAAGCATCACGTGGACTCTACCCTGCCCCTCTGAAAATACTTGAAGTAGTAAGAACAGGAGTGGACAAGGGTCCTGTAGCCGGTTATGAAGCAGAAGCACAAGGCTTTGGTGAACTGGCTATGACTCCTCAGAGCAAAGGGTTGATTGGTTTATTCCGTGGACAAACAGAATGTAAGAAGAATAGGTTCGGAAAATCCCAAGTTGATGTAAAAACGATTGGTGTTCTTGGTGCCGGTTTGATGGGAGCAGGTATAGTGCAAGTGTCAATCGACAAAGGCTACAATGTCGTAATGAAAGACGCCACTAATGCAGGCTTGTATCGCGGCGTTGGACAAATTCAAACCGGACTAAGTAATGCTGTGAAACGCAAAAGAATTTCATCTCTACAAAAGGATAAGTACTTATCGAATCTACTTCCTACATTGGACTATGCCAAGTTCAAAAATGCGGATTGTGTAATTGAAGCCGTTTTCGAGGATATTAACATCAAGCACAAAGTTATCAAAGAGTTAGAGGCTGTAGTGCCGAAACATTGTATTATAGCTACAAACACTAGTGCTATACCTATTACAAAAATCGCAGCTGGCAGCAGTCGTCCTGATAAAGTAATTGGCATGCATTACTTCTCACCAGTTGACAAGATGCAATTGCTAGAAATTATTAAGCACCCCGGCACCAGCGACGATACATCGGCAGCCGCTGTGGCCGTAGGTCTACGACAAGGAAAGGTCGTCATCACGGTGGGGGATGGGCCTGGTTTTTACACCACACGTATCCTTTCTACTATGCTTAGTGAAGCAGTAAGACTGCTACAAGAGGGTGTTGATCCTAAAGAACTGGATAACATGACAAAGCAATTTGGTTTCCCTGTCGGCGCTGCTACATTGGCTGACGAAGTTGGTATCGATGTCGGCTCTCACATCGCTGTGGATCTAGCCAAGGCCTTTGGTGATCGCATTAGCGGAGGAAACTTGGGTATCATGCAGGACTTAGTGCAAGCTGGATACATGGGACGCAAGTCCGGCAAAGGTTTCTATGTGTATGAAAAAGGCTCTAAAAACAGGGAAGTAAACCATGGTGCTATTAAAATCCTGAAAGACAAGTATGCATTAGAACCTCGTGGTGCTAATACTGTTGAAGACCAACAACTGCGAATGGTTTCCAGATTTGTAAATGAAGCAGTGTTATCATTGGAAGAAAAAATTCTACACAACCCCCTTGAAGGTGATGTTGGTGCGGTCTTTGGTCTCGGCTTCCCTCCATTTACTGGTGGACCTTTCAGATGGGTGGATCAATATGGCGCTGATAAATTAGTAAAGAAAATGGAGGAATACCAGGGTCTTTATGGCGCTCCATTCAAACCAGCTCAAACCCTTGTAGATATGGCCAAAGACACAAGTAAAAAATTCTATAAGAACTGA
Protein
MANSKILSALKILRSRKELFISGVHSRKYAVPASQVHTKCKLVNGVYVVTLDSPNVKVNSLNTQVMEEVSNIVNEIETNSGIEAAVIISGKPGCFIAGADISMIENCKTKEEVVSLSKRGHEIFRRIEQSRKPYIAAIQGSCLGGGLETALACKYRIAVKDSKTGFGLPEVMLGLLPGGGGTQRLPALTSIPTTLDLALTGKTVKADKAKKLGIVDLLVSPLGPGLGQPEENTAKYLETVAIQIAKDIAAGKIIVDRSKKGLVQKITAIVMQWEFVKNIIFKKAKEQVMKASRGLYPAPLKILEVVRTGVDKGPVAGYEAEAQGFGELAMTPQSKGLIGLFRGQTECKKNRFGKSQVDVKTIGVLGAGLMGAGIVQVSIDKGYNVVMKDATNAGLYRGVGQIQTGLSNAVKRKRISSLQKDKYLSNLLPTLDYAKFKNADCVIEAVFEDINIKHKVIKELEAVVPKHCIIATNTSAIPITKIAAGSSRPDKVIGMHYFSPVDKMQLLEIIKHPGTSDDTSAAAVAVGLRQGKVVITVGDGPGFYTTRILSTMLSEAVRLLQEGVDPKELDNMTKQFGFPVGAATLADEVGIDVGSHIAVDLAKAFGDRISGGNLGIMQDLVQAGYMGRKSGKGFYVYEKGSKNREVNHGAIKILKDKYALEPRGANTVEDQQLRMVSRFVNEAVLSLEEKILHNPLEGDVGAVFGLGFPPFTGGPFRWVDQYGADKLVKKMEEYQGLYGAPFKPAQTLVDMAKDTSKKFYKN
Summary
Similarity
Belongs to the enoyl-CoA hydratase/isomerase family.
Uniprot
Q2F686
H9J0D1
A0A194PLY5
A0A068FJG9
A0A212FAS2
A0A2H1WAM2
+ More
A0A194QNG0 S4PXY5 A0A182FNA1 A0A2M4BEQ5 A0A2M4BFA5 U5EW00 A0A2M3ZFI0 A0A2M3ZFG6 A0A2M4A1V6 W5JCS5 A0A2M4A1U5 A0A084VI50 A0A182M3A5 A0A182W7P6 A0A182XZN5 A0A182JSD1 A0A182R0N7 Q16TS7 A0A182VD20 B0XLF3 A0A182L2K5 Q7Q3A1 A0A182IEF4 A0A182RU85 A0A182PJP5 A0A182TFW3 A0A182JE28 Q1DGR4 A0A1B0CVB3 A0A182MZA7 T1DQ42 A0A182H6J0 A0A182X1H4 A0A1Q3FDZ4 A0A1L8DYL3 A0A1L8DYI3 A0A3B0KKL1 B4G819 Q29NA3 B4M9G5 B4KH24 B4MTL1 B3MUW0 B3N8F4 B4NY54 Q9V397 A0A1A9YAT6 A0A1A9W002 Q8IPE8 B4HYZ1 A0A1B0B7H6 B4Q7T9 D3TSD9 A0A1B0A4X2 A0A0M4ESC9 A0A1I8MXU8 A0A1W4WA91 T1PFH4 A0A1A9UCN6 A0A1I8P083 A0A0L0C606 B4JCF5 A0A1Y1NJQ7 A0A0T6AT59 A0A336KZW8 A0A1B0FLJ3 N6UKT1 D6WNJ5 A0A232F6Y9 K7IQV3 J3JZ20 A0A182S6H2 E1ZXM8 N6UN62 E2C2R4 A0A026WF63 A0A310SB78 A0A0A1XRE1 A0A1J1IJI3 A0A0M9A3C6 A0A195FVV8 A0A146LFU0 A0A158NLR9 A0A0K8SLU2 A0A0A9Y8J7 A0A0C9RF77 A0A067REB8 A0A195CEJ5 A0A151WN20 A0A1B6DDQ1 A0A195DSE3 A0A069DWP2 A0A224X7Z8 A0A034VAS8
A0A194QNG0 S4PXY5 A0A182FNA1 A0A2M4BEQ5 A0A2M4BFA5 U5EW00 A0A2M3ZFI0 A0A2M3ZFG6 A0A2M4A1V6 W5JCS5 A0A2M4A1U5 A0A084VI50 A0A182M3A5 A0A182W7P6 A0A182XZN5 A0A182JSD1 A0A182R0N7 Q16TS7 A0A182VD20 B0XLF3 A0A182L2K5 Q7Q3A1 A0A182IEF4 A0A182RU85 A0A182PJP5 A0A182TFW3 A0A182JE28 Q1DGR4 A0A1B0CVB3 A0A182MZA7 T1DQ42 A0A182H6J0 A0A182X1H4 A0A1Q3FDZ4 A0A1L8DYL3 A0A1L8DYI3 A0A3B0KKL1 B4G819 Q29NA3 B4M9G5 B4KH24 B4MTL1 B3MUW0 B3N8F4 B4NY54 Q9V397 A0A1A9YAT6 A0A1A9W002 Q8IPE8 B4HYZ1 A0A1B0B7H6 B4Q7T9 D3TSD9 A0A1B0A4X2 A0A0M4ESC9 A0A1I8MXU8 A0A1W4WA91 T1PFH4 A0A1A9UCN6 A0A1I8P083 A0A0L0C606 B4JCF5 A0A1Y1NJQ7 A0A0T6AT59 A0A336KZW8 A0A1B0FLJ3 N6UKT1 D6WNJ5 A0A232F6Y9 K7IQV3 J3JZ20 A0A182S6H2 E1ZXM8 N6UN62 E2C2R4 A0A026WF63 A0A310SB78 A0A0A1XRE1 A0A1J1IJI3 A0A0M9A3C6 A0A195FVV8 A0A146LFU0 A0A158NLR9 A0A0K8SLU2 A0A0A9Y8J7 A0A0C9RF77 A0A067REB8 A0A195CEJ5 A0A151WN20 A0A1B6DDQ1 A0A195DSE3 A0A069DWP2 A0A224X7Z8 A0A034VAS8
Pubmed
19121390
26354079
26385554
22118469
23622113
20920257
+ More
23761445 24438588 25244985 17510324 20966253 12364791 14747013 17210077 26483478 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20353571 25315136 26108605 28004739 23537049 18362917 19820115 28648823 20075255 22516182 20798317 24508170 30249741 25830018 26823975 21347285 25401762 24845553 26334808 25348373
23761445 24438588 25244985 17510324 20966253 12364791 14747013 17210077 26483478 17994087 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 20353571 25315136 26108605 28004739 23537049 18362917 19820115 28648823 20075255 22516182 20798317 24508170 30249741 25830018 26823975 21347285 25401762 24845553 26334808 25348373
EMBL
DQ311186
ABD36131.1
BABH01010547
BABH01010548
KQ459601
KPI93749.1
+ More
KJ622097 AID66687.1 AGBW02009433 OWR50827.1 ODYU01007388 SOQ50103.1 KQ461196 KPJ06510.1 GAIX01004228 JAA88332.1 GGFJ01002356 MBW51497.1 GGFJ01002357 MBW51498.1 GANO01000770 JAB59101.1 GGFM01006565 MBW27316.1 GGFM01006543 MBW27294.1 GGFK01001446 MBW34767.1 ADMH02001533 ETN62152.1 GGFK01001404 MBW34725.1 ATLV01013284 KE524849 KFB37644.1 AXCM01004662 AXCN02002070 CH477641 EAT37915.1 EAT37916.1 DS234448 EDS34040.1 AAAB01008964 EAA12305.4 APCN01005201 CH900762 EAT32343.1 AJWK01030388 GAMD01001400 JAB00191.1 JXUM01114162 JXUM01114163 KQ565759 KXJ70695.1 GFDL01009278 JAV25767.1 GFDF01002560 JAV11524.1 GFDF01002561 JAV11523.1 OUUW01000010 SPP85691.1 CH479180 EDW28499.1 CH379060 EAL33439.2 CH940654 EDW57841.1 CH933807 EDW12235.1 CH963852 EDW75450.1 CH902624 EDV33025.2 CH954177 EDV58377.1 CM000157 EDW88656.1 AF181648 AE014134 AAD55434.1 AAF52789.1 AAN10697.1 CH480818 EDW52271.1 JXJN01009667 CM000361 CM002910 EDX04371.1 KMY89287.1 EZ424341 ADD20617.1 CP012523 ALC40225.1 KA647542 AFP62171.1 JRES01000864 KNC27681.1 CH916368 EDW03109.1 GEZM01001140 JAV98121.1 LJIG01022873 KRT78289.1 UFQS01000186 UFQS01001254 UFQT01000186 UFQT01001254 SSX00923.1 SSX09946.1 SSX21303.1 CCAG010008497 APGK01029294 APGK01029295 KB740731 KB632226 ENN79292.1 ERL90303.1 KQ971343 EFA04382.1 NNAY01000802 OXU26435.1 BT128501 AEE63458.1 GL435067 EFN74066.1 APGK01025474 APGK01025475 KB740591 ENN80162.1 GL452203 EFN77749.1 KK107238 QOIP01000007 EZA54760.1 RLU20785.1 KQ765234 OAD54016.1 GBXI01014555 GBXI01001154 JAC99736.1 JAD13138.1 CVRI01000052 CRK99700.1 KQ435748 KOX76405.1 KQ981219 KYN44447.1 GDHC01011565 JAQ07064.1 ADTU01019758 GBRD01011538 JAG54286.1 GBHO01014212 JAG29392.1 GBYB01011752 JAG81519.1 KK852514 KDR22191.1 KQ977957 KYM98488.1 KQ982919 KYQ49269.1 GEDC01013467 JAS23831.1 KQ980489 KYN15840.1 GBGD01000579 JAC88310.1 GFTR01007950 JAW08476.1 GAKP01018542 GAKP01018541 JAC40411.1
KJ622097 AID66687.1 AGBW02009433 OWR50827.1 ODYU01007388 SOQ50103.1 KQ461196 KPJ06510.1 GAIX01004228 JAA88332.1 GGFJ01002356 MBW51497.1 GGFJ01002357 MBW51498.1 GANO01000770 JAB59101.1 GGFM01006565 MBW27316.1 GGFM01006543 MBW27294.1 GGFK01001446 MBW34767.1 ADMH02001533 ETN62152.1 GGFK01001404 MBW34725.1 ATLV01013284 KE524849 KFB37644.1 AXCM01004662 AXCN02002070 CH477641 EAT37915.1 EAT37916.1 DS234448 EDS34040.1 AAAB01008964 EAA12305.4 APCN01005201 CH900762 EAT32343.1 AJWK01030388 GAMD01001400 JAB00191.1 JXUM01114162 JXUM01114163 KQ565759 KXJ70695.1 GFDL01009278 JAV25767.1 GFDF01002560 JAV11524.1 GFDF01002561 JAV11523.1 OUUW01000010 SPP85691.1 CH479180 EDW28499.1 CH379060 EAL33439.2 CH940654 EDW57841.1 CH933807 EDW12235.1 CH963852 EDW75450.1 CH902624 EDV33025.2 CH954177 EDV58377.1 CM000157 EDW88656.1 AF181648 AE014134 AAD55434.1 AAF52789.1 AAN10697.1 CH480818 EDW52271.1 JXJN01009667 CM000361 CM002910 EDX04371.1 KMY89287.1 EZ424341 ADD20617.1 CP012523 ALC40225.1 KA647542 AFP62171.1 JRES01000864 KNC27681.1 CH916368 EDW03109.1 GEZM01001140 JAV98121.1 LJIG01022873 KRT78289.1 UFQS01000186 UFQS01001254 UFQT01000186 UFQT01001254 SSX00923.1 SSX09946.1 SSX21303.1 CCAG010008497 APGK01029294 APGK01029295 KB740731 KB632226 ENN79292.1 ERL90303.1 KQ971343 EFA04382.1 NNAY01000802 OXU26435.1 BT128501 AEE63458.1 GL435067 EFN74066.1 APGK01025474 APGK01025475 KB740591 ENN80162.1 GL452203 EFN77749.1 KK107238 QOIP01000007 EZA54760.1 RLU20785.1 KQ765234 OAD54016.1 GBXI01014555 GBXI01001154 JAC99736.1 JAD13138.1 CVRI01000052 CRK99700.1 KQ435748 KOX76405.1 KQ981219 KYN44447.1 GDHC01011565 JAQ07064.1 ADTU01019758 GBRD01011538 JAG54286.1 GBHO01014212 JAG29392.1 GBYB01011752 JAG81519.1 KK852514 KDR22191.1 KQ977957 KYM98488.1 KQ982919 KYQ49269.1 GEDC01013467 JAS23831.1 KQ980489 KYN15840.1 GBGD01000579 JAC88310.1 GFTR01007950 JAW08476.1 GAKP01018542 GAKP01018541 JAC40411.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000053240
UP000069272
UP000000673
+ More
UP000030765 UP000075883 UP000075920 UP000076408 UP000075881 UP000075886 UP000008820 UP000075903 UP000002320 UP000075882 UP000007062 UP000075840 UP000075900 UP000075885 UP000075902 UP000075880 UP000092461 UP000075884 UP000069940 UP000249989 UP000076407 UP000268350 UP000008744 UP000001819 UP000008792 UP000009192 UP000007798 UP000007801 UP000008711 UP000002282 UP000000803 UP000092443 UP000091820 UP000001292 UP000092460 UP000000304 UP000092445 UP000092553 UP000095301 UP000192221 UP000078200 UP000095300 UP000037069 UP000001070 UP000092444 UP000019118 UP000030742 UP000007266 UP000215335 UP000002358 UP000075901 UP000000311 UP000008237 UP000053097 UP000279307 UP000183832 UP000053105 UP000078541 UP000005205 UP000027135 UP000078542 UP000075809 UP000078492
UP000030765 UP000075883 UP000075920 UP000076408 UP000075881 UP000075886 UP000008820 UP000075903 UP000002320 UP000075882 UP000007062 UP000075840 UP000075900 UP000075885 UP000075902 UP000075880 UP000092461 UP000075884 UP000069940 UP000249989 UP000076407 UP000268350 UP000008744 UP000001819 UP000008792 UP000009192 UP000007798 UP000007801 UP000008711 UP000002282 UP000000803 UP000092443 UP000091820 UP000001292 UP000092460 UP000000304 UP000092445 UP000092553 UP000095301 UP000192221 UP000078200 UP000095300 UP000037069 UP000001070 UP000092444 UP000019118 UP000030742 UP000007266 UP000215335 UP000002358 UP000075901 UP000000311 UP000008237 UP000053097 UP000279307 UP000183832 UP000053105 UP000078541 UP000005205 UP000027135 UP000078542 UP000075809 UP000078492
Interpro
IPR029045
ClpP/crotonase-like_dom_sf
+ More
IPR006176 3-OHacyl-CoA_DH_NAD-bd
IPR012803 Fa_ox_alpha_mit
IPR008927 6-PGluconate_DH-like_C_sf
IPR006108 3HC_DH_C
IPR036291 NAD(P)-bd_dom_sf
IPR001753 Enoyl-CoA_hydra/iso
IPR018376 Enoyl-CoA_hyd/isom_CS
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR020846 MFS_dom
IPR006623 THEG
IPR006180 3-OHacyl-CoA_DH_CS
IPR006176 3-OHacyl-CoA_DH_NAD-bd
IPR012803 Fa_ox_alpha_mit
IPR008927 6-PGluconate_DH-like_C_sf
IPR006108 3HC_DH_C
IPR036291 NAD(P)-bd_dom_sf
IPR001753 Enoyl-CoA_hydra/iso
IPR018376 Enoyl-CoA_hyd/isom_CS
IPR036259 MFS_trans_sf
IPR011701 MFS
IPR020846 MFS_dom
IPR006623 THEG
IPR006180 3-OHacyl-CoA_DH_CS
CDD
ProteinModelPortal
Q2F686
H9J0D1
A0A194PLY5
A0A068FJG9
A0A212FAS2
A0A2H1WAM2
+ More
A0A194QNG0 S4PXY5 A0A182FNA1 A0A2M4BEQ5 A0A2M4BFA5 U5EW00 A0A2M3ZFI0 A0A2M3ZFG6 A0A2M4A1V6 W5JCS5 A0A2M4A1U5 A0A084VI50 A0A182M3A5 A0A182W7P6 A0A182XZN5 A0A182JSD1 A0A182R0N7 Q16TS7 A0A182VD20 B0XLF3 A0A182L2K5 Q7Q3A1 A0A182IEF4 A0A182RU85 A0A182PJP5 A0A182TFW3 A0A182JE28 Q1DGR4 A0A1B0CVB3 A0A182MZA7 T1DQ42 A0A182H6J0 A0A182X1H4 A0A1Q3FDZ4 A0A1L8DYL3 A0A1L8DYI3 A0A3B0KKL1 B4G819 Q29NA3 B4M9G5 B4KH24 B4MTL1 B3MUW0 B3N8F4 B4NY54 Q9V397 A0A1A9YAT6 A0A1A9W002 Q8IPE8 B4HYZ1 A0A1B0B7H6 B4Q7T9 D3TSD9 A0A1B0A4X2 A0A0M4ESC9 A0A1I8MXU8 A0A1W4WA91 T1PFH4 A0A1A9UCN6 A0A1I8P083 A0A0L0C606 B4JCF5 A0A1Y1NJQ7 A0A0T6AT59 A0A336KZW8 A0A1B0FLJ3 N6UKT1 D6WNJ5 A0A232F6Y9 K7IQV3 J3JZ20 A0A182S6H2 E1ZXM8 N6UN62 E2C2R4 A0A026WF63 A0A310SB78 A0A0A1XRE1 A0A1J1IJI3 A0A0M9A3C6 A0A195FVV8 A0A146LFU0 A0A158NLR9 A0A0K8SLU2 A0A0A9Y8J7 A0A0C9RF77 A0A067REB8 A0A195CEJ5 A0A151WN20 A0A1B6DDQ1 A0A195DSE3 A0A069DWP2 A0A224X7Z8 A0A034VAS8
A0A194QNG0 S4PXY5 A0A182FNA1 A0A2M4BEQ5 A0A2M4BFA5 U5EW00 A0A2M3ZFI0 A0A2M3ZFG6 A0A2M4A1V6 W5JCS5 A0A2M4A1U5 A0A084VI50 A0A182M3A5 A0A182W7P6 A0A182XZN5 A0A182JSD1 A0A182R0N7 Q16TS7 A0A182VD20 B0XLF3 A0A182L2K5 Q7Q3A1 A0A182IEF4 A0A182RU85 A0A182PJP5 A0A182TFW3 A0A182JE28 Q1DGR4 A0A1B0CVB3 A0A182MZA7 T1DQ42 A0A182H6J0 A0A182X1H4 A0A1Q3FDZ4 A0A1L8DYL3 A0A1L8DYI3 A0A3B0KKL1 B4G819 Q29NA3 B4M9G5 B4KH24 B4MTL1 B3MUW0 B3N8F4 B4NY54 Q9V397 A0A1A9YAT6 A0A1A9W002 Q8IPE8 B4HYZ1 A0A1B0B7H6 B4Q7T9 D3TSD9 A0A1B0A4X2 A0A0M4ESC9 A0A1I8MXU8 A0A1W4WA91 T1PFH4 A0A1A9UCN6 A0A1I8P083 A0A0L0C606 B4JCF5 A0A1Y1NJQ7 A0A0T6AT59 A0A336KZW8 A0A1B0FLJ3 N6UKT1 D6WNJ5 A0A232F6Y9 K7IQV3 J3JZ20 A0A182S6H2 E1ZXM8 N6UN62 E2C2R4 A0A026WF63 A0A310SB78 A0A0A1XRE1 A0A1J1IJI3 A0A0M9A3C6 A0A195FVV8 A0A146LFU0 A0A158NLR9 A0A0K8SLU2 A0A0A9Y8J7 A0A0C9RF77 A0A067REB8 A0A195CEJ5 A0A151WN20 A0A1B6DDQ1 A0A195DSE3 A0A069DWP2 A0A224X7Z8 A0A034VAS8
PDB
5ZRV
E-value=0,
Score=2066
Ontologies
PATHWAY
00062
Fatty acid elongation - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
00071 Fatty acid degradation - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00410 beta-Alanine metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
GO
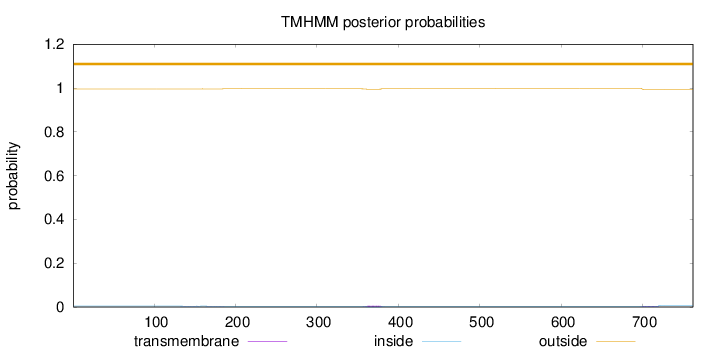
Topology
Length:
762
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.227390000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00442
outside
1 - 762
Population Genetic Test Statistics
Pi
213.028583
Theta
202.937548
Tajima's D
-2.023671
CLR
0.517831
CSRT
0.0153492325383731
Interpretation
Uncertain
