Pre Gene Modal
BGIBMGA003207
Annotation
PREDICTED:_negative_elongation_factor_E_[Amyelois_transitella]
Full name
Negative elongation factor E
Location in the cell
Nuclear Reliability : 4.147
Sequence
CDS
ATGGTCTATTTACATTTTCCTTCGAACCTAACCGAGGAAGAACTAATGCTACAAGCTAAATACCAGAAACTAAAAAAGAAGAAGAAAGCTTTGCAAGCCCTTAAGGCACCAAAACCCGAACCAGAAAAGCCAATTTTACCAAAAAGGCCTACAGAAGCCAGAGATGCCAGGGAAATAGCTAGAAAATTGCTGAAAAGTGGTGCTATTCAAGCTATTAAAAGTCCTCCAACTCAGCCACAGAACACCACTTTCAAAAAACCACGTGCAGGTCGAGAAAGGAAATTAAGTGGTTCTGCTGGAGTACCTGGAGGTGTTGCTTCATATCAACCATTCAGTGCTTCACAAGAACCTCCACCTCAAGAAGAGAAACAAACTCCAAGAGTTAAATATCTTTATGACTCTTTTGTCACTGCCAGAGAAAAGGACGACAAAGGCTCACGCACACCCAATCCAGGTGGCAGTACACCGTCTCAATCAGATGGTCGCAGTTGTACTTTACAATTGGTCGGTCAACGGATATCTGAAGAGATATTGAGGAGACGCCTGGCTACTTTTGAGCCCTTTGCATTCATGCCAAAAGATGACAAGGCTTTCTTAACCTTCGATAGTCCTGACATTGCGTCGCACGCTTTGTCGGCTCTTGAAGGCAACGAGGAGGGCTGGCGTATAAACATCGCGAGAAGACCTCCGCAGCAACAAGGGACTTGGAGTGCGTCGTCTTCACCACGAAATCCACAACCGCCTCCCAAGGACTTGAAACGTACATTATTGACTTATGATGATATATTTGATTAA
Protein
MVYLHFPSNLTEEELMLQAKYQKLKKKKKALQALKAPKPEPEKPILPKRPTEARDAREIARKLLKSGAIQAIKSPPTQPQNTTFKKPRAGRERKLSGSAGVPGGVASYQPFSASQEPPPQEEKQTPRVKYLYDSFVTAREKDDKGSRTPNPGGSTPSQSDGRSCTLQLVGQRISEEILRRRLATFEPFAFMPKDDKAFLTFDSPDIASHALSALEGNEEGWRINIARRPPQQQGTWSASSSPRNPQPPPKDLKRTLLTYDDIFD
Summary
Description
Essential component of the NELF complex, a complex that negatively regulates the elongation of transcription by RNA polymerase II by RNA polymerase II. The NELF complex, which acts via an association with the DSIF complex, causes transcriptional pausing.
Essential component of the NELF complex, a complex that negatively regulates the elongation of transcription by RNA polymerase II by RNA polymerase II. The NELF complex, which acts via an association with the DSIF complex, causes transcriptional pausing (By similarity).
Essential component of the NELF complex, a complex that negatively regulates the elongation of transcription by RNA polymerase II by RNA polymerase II. The NELF complex, which acts via an association with the DSIF complex, causes transcriptional pausing (By similarity).
Subunit
Component of the NELF complex, which is at least composed of TH1/Nelf-D and Nelf-E.
Component of the NELF complex, which is at least composed of TH1/NELF-D and NELF-E.
Component of the NELF complex, which is at least composed of TH1/NELF-D and NELF-E.
Similarity
Belongs to the RRM NELF-E family.
Keywords
Chromosome
Complete proteome
Nucleus
Reference proteome
Repressor
RNA-binding
Transcription
Transcription regulation
Feature
chain Negative elongation factor E
Uniprot
H9J120
A0A212EHU8
A0A2H1VZM0
A0A2A4K4E6
A0A194PL88
I4DRH2
+ More
A0A0L7L4E7 A0A194QME1 S4PKE8 A0A1E1WBN8 A0A1B6M8G1 A0A1B6G9Y2 A0A067R779 V5FXT6 A0A088A9Q8 D6WEE5 N6UEI7 A0A1Y1LEN3 A0A232FDW5 A0A310S6S8 A0A0L7QX51 A0A182YKZ2 E0W480 A0A182LLJ2 Q7Q5S3 A0A2A3EP91 A0A1L8DSL7 Q0IG90 A0A2M4AUP3 W5JIR8 A0A154P8M4 A0A1B6EGY2 A0A2M3ZCG7 A0A2J7RG74 E9HI81 U5ENW6 W8B6I8 A0A182MIL1 A0A0P6DW17 A0A2M4BVC0 A0A182I161 A0A1A9UJC6 A0A182VN84 A0A1W4WJN1 A0A1A9YN44 A0A1B0BHX8 A0A182WLS0 A0A182UE00 B4KZ57 A0A1A9Z2R2 E2AJZ5 A0A151WWJ0 E2BRF5 B3NBR3 A0A023EMA8 A0A0J7K8A0 B4PG02 A0A2M4BVD4 A0A2M4BWB4 A0A182JSS4 K7J8E4 A0A182PL93 A0A182FHR0 A0A1W4US37 A0A1S4F3Y9 A0A1A9WIA1 P92204 A0A0J9RT11 B4HJZ8 A0A158NNJ0 A0A195B2P9 A0A195DKM2 A0A0N0U3M1 A0A151I9U1 B4IWM1 A0A195F6M1 B4LEK7 A0A1S4EPU6 A0A026WC07 A0A182H4S0 F4WQF1 A0A336MND3 A0A0N8AZR8 A0A336M931 Q95ZE9 A0A0K8UF00 A0A034VPP1 A0A0P6AG97 A0A1I8MR11 B3M688 A0A182N947 B4MXU2 A0A1I8PRN4 A0A182QLE9 A0A3B0JBI9 A0A182X8L1 Q29D00 B4H9I0 A0A2P2IAF6
A0A0L7L4E7 A0A194QME1 S4PKE8 A0A1E1WBN8 A0A1B6M8G1 A0A1B6G9Y2 A0A067R779 V5FXT6 A0A088A9Q8 D6WEE5 N6UEI7 A0A1Y1LEN3 A0A232FDW5 A0A310S6S8 A0A0L7QX51 A0A182YKZ2 E0W480 A0A182LLJ2 Q7Q5S3 A0A2A3EP91 A0A1L8DSL7 Q0IG90 A0A2M4AUP3 W5JIR8 A0A154P8M4 A0A1B6EGY2 A0A2M3ZCG7 A0A2J7RG74 E9HI81 U5ENW6 W8B6I8 A0A182MIL1 A0A0P6DW17 A0A2M4BVC0 A0A182I161 A0A1A9UJC6 A0A182VN84 A0A1W4WJN1 A0A1A9YN44 A0A1B0BHX8 A0A182WLS0 A0A182UE00 B4KZ57 A0A1A9Z2R2 E2AJZ5 A0A151WWJ0 E2BRF5 B3NBR3 A0A023EMA8 A0A0J7K8A0 B4PG02 A0A2M4BVD4 A0A2M4BWB4 A0A182JSS4 K7J8E4 A0A182PL93 A0A182FHR0 A0A1W4US37 A0A1S4F3Y9 A0A1A9WIA1 P92204 A0A0J9RT11 B4HJZ8 A0A158NNJ0 A0A195B2P9 A0A195DKM2 A0A0N0U3M1 A0A151I9U1 B4IWM1 A0A195F6M1 B4LEK7 A0A1S4EPU6 A0A026WC07 A0A182H4S0 F4WQF1 A0A336MND3 A0A0N8AZR8 A0A336M931 Q95ZE9 A0A0K8UF00 A0A034VPP1 A0A0P6AG97 A0A1I8MR11 B3M688 A0A182N947 B4MXU2 A0A1I8PRN4 A0A182QLE9 A0A3B0JBI9 A0A182X8L1 Q29D00 B4H9I0 A0A2P2IAF6
Pubmed
19121390
22118469
26354079
22651552
26227816
23622113
+ More
24845553 18362917 19820115 23537049 28004739 28648823 25244985 20566863 20966253 12364791 17510324 20920257 23761445 21292972 24495485 17994087 20798317 24945155 26483478 17550304 20075255 9236770 10731132 12537572 12537569 12782658 22936249 21347285 24508170 30249741 21719571 11814692 25348373 25315136 15632085
24845553 18362917 19820115 23537049 28004739 28648823 25244985 20566863 20966253 12364791 17510324 20920257 23761445 21292972 24495485 17994087 20798317 24945155 26483478 17550304 20075255 9236770 10731132 12537572 12537569 12782658 22936249 21347285 24508170 30249741 21719571 11814692 25348373 25315136 15632085
EMBL
BABH01010587
AGBW02014799
OWR41055.1
ODYU01005437
SOQ46281.1
NWSH01000140
+ More
PCG79131.1 KQ459601 KPI93763.1 AK405118 BAM20512.1 JTDY01002995 KOB70352.1 KQ461196 KPJ06524.1 GAIX01001036 JAA91524.1 GDQN01006646 JAT84408.1 GEBQ01023873 GEBQ01007761 JAT16104.1 JAT32216.1 GECZ01010533 JAS59236.1 KK852694 KDR18301.1 GALX01005986 JAB62480.1 KQ971318 EFA00397.1 APGK01025467 KB740591 KB632226 ENN80155.1 ERL90296.1 GEZM01057940 JAV72084.1 NNAY01000392 OXU28693.1 KQ767283 OAD53454.1 KQ414705 KOC63119.1 DS235886 EEB20436.1 AAAB01008960 EAA10770.4 KZ288212 PBC32851.1 GFDF01004769 JAV09315.1 CH477261 EAT45667.1 GGFK01011184 MBW44505.1 ADMH02001054 ETN64277.1 KQ434846 KZC08269.1 GEDC01027159 GEDC01023196 GEDC01019531 GEDC01003222 GEDC01000102 JAS10139.1 JAS14102.1 JAS17767.1 JAS34076.1 JAS37196.1 GGFM01005424 MBW26175.1 NEVH01004403 PNF39839.1 GL732653 EFX68534.1 GANO01000418 JAB59453.1 GAMC01009755 JAB96800.1 AXCM01000856 GDIP01158400 GDIQ01071421 LRGB01003128 JAJ65002.1 JAN23316.1 KZS04155.1 GGFJ01007876 MBW57017.1 APCN01000031 JXJN01014624 CH933809 EDW17854.1 GL440121 EFN66240.1 KQ982691 KYQ52274.1 GL449962 EFN81733.1 CH954178 EDV50659.1 JXUM01009569 GAPW01003162 KQ560286 JAC10436.1 KXJ83270.1 LBMM01011969 KMQ86514.1 CM000159 EDW93146.1 GGFJ01007878 MBW57019.1 GGFJ01007877 MBW57018.1 Y10015 Y10016 AE014296 AY071061 CM002912 KMY98424.1 CH480815 EDW40734.1 ADTU01021611 KQ976641 KYM78748.1 KQ980765 KYN13428.1 KQ435891 KOX69424.1 KQ978273 KYM95777.1 CH916366 EDV96247.1 KQ981744 KYN36245.1 CH940647 EDW69092.1 KK107288 QOIP01000013 EZA53508.1 RLU15488.1 JXUM01025526 KQ560725 KXJ81123.1 GL888273 EGI63551.1 UFQT01001837 SSX31964.1 GDIQ01226915 JAK24810.1 UFQT01000264 SSX22558.1 AJ306692 GDHF01029381 GDHF01027141 GDHF01017011 GDHF01006976 JAI22933.1 JAI25173.1 JAI35303.1 JAI45338.1 GAKP01015182 JAC43770.1 GDIP01031154 JAM72561.1 CH902618 EDV39708.1 CH963876 EDW76861.1 AXCN02000488 OUUW01000002 SPP77402.1 CH379070 EAL30614.1 CH479229 EDW36456.1 IACF01005414 LAB70999.1
PCG79131.1 KQ459601 KPI93763.1 AK405118 BAM20512.1 JTDY01002995 KOB70352.1 KQ461196 KPJ06524.1 GAIX01001036 JAA91524.1 GDQN01006646 JAT84408.1 GEBQ01023873 GEBQ01007761 JAT16104.1 JAT32216.1 GECZ01010533 JAS59236.1 KK852694 KDR18301.1 GALX01005986 JAB62480.1 KQ971318 EFA00397.1 APGK01025467 KB740591 KB632226 ENN80155.1 ERL90296.1 GEZM01057940 JAV72084.1 NNAY01000392 OXU28693.1 KQ767283 OAD53454.1 KQ414705 KOC63119.1 DS235886 EEB20436.1 AAAB01008960 EAA10770.4 KZ288212 PBC32851.1 GFDF01004769 JAV09315.1 CH477261 EAT45667.1 GGFK01011184 MBW44505.1 ADMH02001054 ETN64277.1 KQ434846 KZC08269.1 GEDC01027159 GEDC01023196 GEDC01019531 GEDC01003222 GEDC01000102 JAS10139.1 JAS14102.1 JAS17767.1 JAS34076.1 JAS37196.1 GGFM01005424 MBW26175.1 NEVH01004403 PNF39839.1 GL732653 EFX68534.1 GANO01000418 JAB59453.1 GAMC01009755 JAB96800.1 AXCM01000856 GDIP01158400 GDIQ01071421 LRGB01003128 JAJ65002.1 JAN23316.1 KZS04155.1 GGFJ01007876 MBW57017.1 APCN01000031 JXJN01014624 CH933809 EDW17854.1 GL440121 EFN66240.1 KQ982691 KYQ52274.1 GL449962 EFN81733.1 CH954178 EDV50659.1 JXUM01009569 GAPW01003162 KQ560286 JAC10436.1 KXJ83270.1 LBMM01011969 KMQ86514.1 CM000159 EDW93146.1 GGFJ01007878 MBW57019.1 GGFJ01007877 MBW57018.1 Y10015 Y10016 AE014296 AY071061 CM002912 KMY98424.1 CH480815 EDW40734.1 ADTU01021611 KQ976641 KYM78748.1 KQ980765 KYN13428.1 KQ435891 KOX69424.1 KQ978273 KYM95777.1 CH916366 EDV96247.1 KQ981744 KYN36245.1 CH940647 EDW69092.1 KK107288 QOIP01000013 EZA53508.1 RLU15488.1 JXUM01025526 KQ560725 KXJ81123.1 GL888273 EGI63551.1 UFQT01001837 SSX31964.1 GDIQ01226915 JAK24810.1 UFQT01000264 SSX22558.1 AJ306692 GDHF01029381 GDHF01027141 GDHF01017011 GDHF01006976 JAI22933.1 JAI25173.1 JAI35303.1 JAI45338.1 GAKP01015182 JAC43770.1 GDIP01031154 JAM72561.1 CH902618 EDV39708.1 CH963876 EDW76861.1 AXCN02000488 OUUW01000002 SPP77402.1 CH379070 EAL30614.1 CH479229 EDW36456.1 IACF01005414 LAB70999.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000037510
UP000053240
+ More
UP000027135 UP000005203 UP000007266 UP000019118 UP000030742 UP000215335 UP000053825 UP000076408 UP000009046 UP000075882 UP000007062 UP000242457 UP000008820 UP000000673 UP000076502 UP000235965 UP000000305 UP000075883 UP000076858 UP000075840 UP000078200 UP000075903 UP000192223 UP000092443 UP000092460 UP000075920 UP000075902 UP000009192 UP000092445 UP000000311 UP000075809 UP000008237 UP000008711 UP000069940 UP000249989 UP000036403 UP000002282 UP000075881 UP000002358 UP000075885 UP000069272 UP000192221 UP000091820 UP000000803 UP000001292 UP000005205 UP000078540 UP000078492 UP000053105 UP000078542 UP000001070 UP000078541 UP000008792 UP000079169 UP000053097 UP000279307 UP000007755 UP000095301 UP000007801 UP000075884 UP000007798 UP000095300 UP000075886 UP000268350 UP000076407 UP000001819 UP000008744
UP000027135 UP000005203 UP000007266 UP000019118 UP000030742 UP000215335 UP000053825 UP000076408 UP000009046 UP000075882 UP000007062 UP000242457 UP000008820 UP000000673 UP000076502 UP000235965 UP000000305 UP000075883 UP000076858 UP000075840 UP000078200 UP000075903 UP000192223 UP000092443 UP000092460 UP000075920 UP000075902 UP000009192 UP000092445 UP000000311 UP000075809 UP000008237 UP000008711 UP000069940 UP000249989 UP000036403 UP000002282 UP000075881 UP000002358 UP000075885 UP000069272 UP000192221 UP000091820 UP000000803 UP000001292 UP000005205 UP000078540 UP000078492 UP000053105 UP000078542 UP000001070 UP000078541 UP000008792 UP000079169 UP000053097 UP000279307 UP000007755 UP000095301 UP000007801 UP000075884 UP000007798 UP000095300 UP000075886 UP000268350 UP000076407 UP000001819 UP000008744
Interpro
Gene 3D
ProteinModelPortal
H9J120
A0A212EHU8
A0A2H1VZM0
A0A2A4K4E6
A0A194PL88
I4DRH2
+ More
A0A0L7L4E7 A0A194QME1 S4PKE8 A0A1E1WBN8 A0A1B6M8G1 A0A1B6G9Y2 A0A067R779 V5FXT6 A0A088A9Q8 D6WEE5 N6UEI7 A0A1Y1LEN3 A0A232FDW5 A0A310S6S8 A0A0L7QX51 A0A182YKZ2 E0W480 A0A182LLJ2 Q7Q5S3 A0A2A3EP91 A0A1L8DSL7 Q0IG90 A0A2M4AUP3 W5JIR8 A0A154P8M4 A0A1B6EGY2 A0A2M3ZCG7 A0A2J7RG74 E9HI81 U5ENW6 W8B6I8 A0A182MIL1 A0A0P6DW17 A0A2M4BVC0 A0A182I161 A0A1A9UJC6 A0A182VN84 A0A1W4WJN1 A0A1A9YN44 A0A1B0BHX8 A0A182WLS0 A0A182UE00 B4KZ57 A0A1A9Z2R2 E2AJZ5 A0A151WWJ0 E2BRF5 B3NBR3 A0A023EMA8 A0A0J7K8A0 B4PG02 A0A2M4BVD4 A0A2M4BWB4 A0A182JSS4 K7J8E4 A0A182PL93 A0A182FHR0 A0A1W4US37 A0A1S4F3Y9 A0A1A9WIA1 P92204 A0A0J9RT11 B4HJZ8 A0A158NNJ0 A0A195B2P9 A0A195DKM2 A0A0N0U3M1 A0A151I9U1 B4IWM1 A0A195F6M1 B4LEK7 A0A1S4EPU6 A0A026WC07 A0A182H4S0 F4WQF1 A0A336MND3 A0A0N8AZR8 A0A336M931 Q95ZE9 A0A0K8UF00 A0A034VPP1 A0A0P6AG97 A0A1I8MR11 B3M688 A0A182N947 B4MXU2 A0A1I8PRN4 A0A182QLE9 A0A3B0JBI9 A0A182X8L1 Q29D00 B4H9I0 A0A2P2IAF6
A0A0L7L4E7 A0A194QME1 S4PKE8 A0A1E1WBN8 A0A1B6M8G1 A0A1B6G9Y2 A0A067R779 V5FXT6 A0A088A9Q8 D6WEE5 N6UEI7 A0A1Y1LEN3 A0A232FDW5 A0A310S6S8 A0A0L7QX51 A0A182YKZ2 E0W480 A0A182LLJ2 Q7Q5S3 A0A2A3EP91 A0A1L8DSL7 Q0IG90 A0A2M4AUP3 W5JIR8 A0A154P8M4 A0A1B6EGY2 A0A2M3ZCG7 A0A2J7RG74 E9HI81 U5ENW6 W8B6I8 A0A182MIL1 A0A0P6DW17 A0A2M4BVC0 A0A182I161 A0A1A9UJC6 A0A182VN84 A0A1W4WJN1 A0A1A9YN44 A0A1B0BHX8 A0A182WLS0 A0A182UE00 B4KZ57 A0A1A9Z2R2 E2AJZ5 A0A151WWJ0 E2BRF5 B3NBR3 A0A023EMA8 A0A0J7K8A0 B4PG02 A0A2M4BVD4 A0A2M4BWB4 A0A182JSS4 K7J8E4 A0A182PL93 A0A182FHR0 A0A1W4US37 A0A1S4F3Y9 A0A1A9WIA1 P92204 A0A0J9RT11 B4HJZ8 A0A158NNJ0 A0A195B2P9 A0A195DKM2 A0A0N0U3M1 A0A151I9U1 B4IWM1 A0A195F6M1 B4LEK7 A0A1S4EPU6 A0A026WC07 A0A182H4S0 F4WQF1 A0A336MND3 A0A0N8AZR8 A0A336M931 Q95ZE9 A0A0K8UF00 A0A034VPP1 A0A0P6AG97 A0A1I8MR11 B3M688 A0A182N947 B4MXU2 A0A1I8PRN4 A0A182QLE9 A0A3B0JBI9 A0A182X8L1 Q29D00 B4H9I0 A0A2P2IAF6
Ontologies
GO
PANTHER
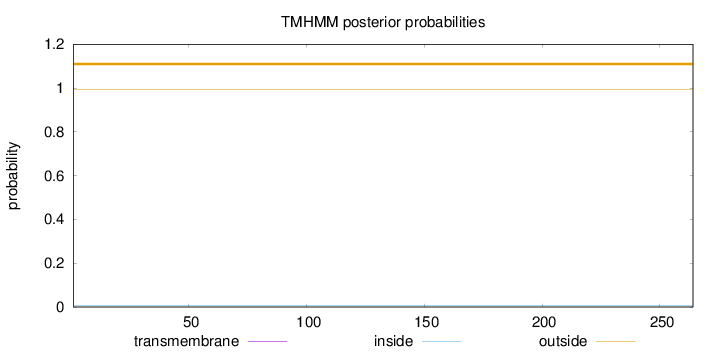
Topology
Subcellular location
Nucleus
Chromosome
Chromosome
Length:
264
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00789
outside
1 - 264
Population Genetic Test Statistics
Pi
49.436336
Theta
6.587306
Tajima's D
-0.764273
CLR
0.711706
CSRT
0.182740862956852
Interpretation
Uncertain
