Gene
KWMTBOMO02034
Pre Gene Modal
BGIBMGA002958
Annotation
PREDICTED:_aromatic-L-amino-acid_decarboxylase-like_[Amyelois_transitella]
Full name
3,4-dihydroxyphenylacetaldehyde synthase
Alternative Name
Alpha-methyldopa hypersensitive protein
Location in the cell
Cytoplasmic Reliability : 3.039
Sequence
CDS
ATGGACGCGAACCAGTTTCGGGAATTCGGCAGAGCAGTCATCGATATGTTGGCGAGCTACGCTGAAAACATAAGGGATTATGATGTTCTGCCGTCTGTTGAACCTGGTTACTTGTTAAGAGCTCTGCCTGAAAGTGCACCCGAACAACCGGAGGATTGGAAAGACATAATGAAAGATTTCAATCAATCAATAATGCCGGGTGTAACACACTGGCAATCTCCGCAGTTCCATGCATTCTATCCTTCTGGCTCCTCATTCGCGAGTATTATAGGAAATATGCTTAGTGACGGCTTGGCGGTTGTGGGATTTAGTTGGATGGCTAGCCCTGCGTGTACAGAACTGGAGGTGGTCACGATGAATTGGCTTGGTAAGCTATTAGATTTACCTGAAGAATTTCTAAATTGCTCTTCGGGTCCCGGAGGCGGAGTTATTCAAGGATCAGCGAGCGAAGCCACCTTAGTTGGATTGCTTGTAGCCAAAGATAAAACTGTTCGCCGTTTCATGAATAATAATCCAGATCTCGATGAAAACGAGATAAAAGCAAAACTTGTAGCTTATACATCCGACCAATGTAACTCGTCAGTAGAAAAAGCCGGCTTACTTGGTTCTATGAAAATGAAACTGTTGAAAGCGGATGCTGATGGATGCCTACGCGGAGAAACATTGAAAAGGGCAATCGAAGAAGATAAGTCGCAAGGACTTATACCCTGCTATGTCGTCGCTAATCTGGGAACAACTGGAACTTGTGCATTCGATCCTTTACATGAATTAGGGCCAATATGTAGTGAGGAAGATATATGGCTTCACGTAGATGCAGCCTATGCGGGAGCAGCATTTTTGTGTCCTGAATACAGACACCTAATGAAAGGTATTGAACATTCTCAATCGTTCGTAACGAATGCACATAAGTGGCTACCGGTTAATTTCGATTGCTCTGCTATGTGGGTTAAAAATGGTTATGATATAACGAGAGCATTCGATGTACAAAGAATTTATTTGGATGATGTAAAAACGACAATCAAGATCCCAGATTACAGACACTGGCAAATGCCACTAGGCCGTCGCTTTAGAGCCTTAAAATTGTGGACAGTTATGAGAATTTATGGTGCTGAAGGTTTGAAAACGCATATCAGACAGCAAATAGAATTAGCACAGTATTTTGCAAAACTGGTACGTGCGGACGAACGTTTTGTGATTGGGCCCGAACCGACTATGGCATTGGTCTGTTTTAGACTGAAAGACGGTGACACAATTACGCGACAATTGTTGGAAAATATAACGCAAAAAAAGAAAGTGTTTATGGTGGCCGGAACGCACAGGGATAGATACGTCATTAGATTCGTGATCTGCTCCCGATTGACTAAGAAGGAAGACGTCGATTACAGCTGGAGCCAAATAAAGAAAGAAACCGACCTCATCTATTCAGATAAAATACACAATAAAGCACAAATACCAGCTCTTGAACAATTCACTTCAAGAGAACTATGCGAAAAATCTAAGTAA
Protein
MDANQFREFGRAVIDMLASYAENIRDYDVLPSVEPGYLLRALPESAPEQPEDWKDIMKDFNQSIMPGVTHWQSPQFHAFYPSGSSFASIIGNMLSDGLAVVGFSWMASPACTELEVVTMNWLGKLLDLPEEFLNCSSGPGGGVIQGSASEATLVGLLVAKDKTVRRFMNNNPDLDENEIKAKLVAYTSDQCNSSVEKAGLLGSMKMKLLKADADGCLRGETLKRAIEEDKSQGLIPCYVVANLGTTGTCAFDPLHELGPICSEEDIWLHVDAAYAGAAFLCPEYRHLMKGIEHSQSFVTNAHKWLPVNFDCSAMWVKNGYDITRAFDVQRIYLDDVKTTIKIPDYRHWQMPLGRRFRALKLWTVMRIYGAEGLKTHIRQQIELAQYFAKLVRADERFVIGPEPTMALVCFRLKDGDTITRQLLENITQKKKVFMVAGTHRDRYVIRFVICSRLTKKEDVDYSWSQIKKETDLIYSDKIHNKAQIPALEQFTSRELCEKSK
Summary
Description
Catalyzes both the decarboxylation and deamination of L-dopa to 3,4-dihydroxylphenylacetaldehyde (DHPAA) (PubMed:21283636). Probably responsible for the protein cross-linking during the development of flexible cuticles (PubMed:21283636). Participates in catecholamine catabolism (PubMed:21283636).
Catalytic Activity
H(+) + H2O + L-dopa + O2 = 3,4-dihydroxyphenylacetaldehyde + CO2 + H2O2 + NH4(+)
Cofactor
pyridoxal 5'-phosphate
Similarity
Belongs to the group II decarboxylase family.
Keywords
Catecholamine metabolism
Complete proteome
Cuticle
Decarboxylase
Lyase
Pyridoxal phosphate
Reference proteome
3D-structure
Alternative splicing
Feature
chain 3,4-dihydroxyphenylacetaldehyde synthase
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J0C2
A0A2A4K5T6
A0A2H1VZS5
A0A194PM00
I4DK34
A0A194QSK8
+ More
A0A212ESM5 A0A0L7K538 A0A1Y1KSC4 A0A1Y1L3F9 A0A0C9QAN0 D6WLR5 A0A2J7RMJ6 N6U414 K7IPX7 A0A2P8ZLM8 A0A154P0I3 A0A2S2QGA6 A0A1B0DQW6 A0A1B0GK69 A0A195EXD1 F4X3J7 A0A182QMB0 A0A2S2NBF5 A0A3L8D9T2 E2BTV0 A0A026VY31 J9JNE6 A0A088A3U2 A0A158NRN0 A0A182R9R3 A0A182KUR6 A0A182X2X5 A0A084VMD9 A0A182UWL6 A0A182I4X9 A0A1S4H0U8 A0A1B6CN44 W5J8H7 A0A182YFF7 A0A195BLR4 A0A182TK73 A0A182NEU9 A0A2H8TM60 A0A182WGY1 A0A182SUA0 A0A232FB20 Q16S21 Q16S22 A0A1B6FZN6 A0A182LXN1 A0A023F2F5 T1HPP9 T1HLH4 E9RJV1 Q6DLV1 A7U8C8 A0A023ET86 A0A023ESV7 B4KKG6 A0A0T6AX31 A0A182JDS8 D6WLA0 B3MNP5 A0A0N0BH44 Q9NL84 A0A1S5QPR3 A0A067RL11 Q16GW0 Q16GW1 Q16895 A0A1Q3FVH7 A0A1J1HJ38 T1DQ67 A0A2A4IYP3 A0A1W4VXM7 J3JUQ3 A0A1D2NMP1 A0A2P8ZLL9 B4N186 A0A1A9V9K7 A0A1A9XGU4 W5J9T6 A0A1I8NE52 A0A1B6LPG7 A0A224XNZ0 A0A182F5H7 A0A0V0G4L0 A0A1L1ZLS9 A0A0Q5VXY7 M4GVV8 C3ZRR8 A0A2H1WW81 A0A1Y1KPJ8 H5V873 P18486 A0A0R1DPK1 A0A220K8N1 C9QP35 A0A3B5KIE6
A0A212ESM5 A0A0L7K538 A0A1Y1KSC4 A0A1Y1L3F9 A0A0C9QAN0 D6WLR5 A0A2J7RMJ6 N6U414 K7IPX7 A0A2P8ZLM8 A0A154P0I3 A0A2S2QGA6 A0A1B0DQW6 A0A1B0GK69 A0A195EXD1 F4X3J7 A0A182QMB0 A0A2S2NBF5 A0A3L8D9T2 E2BTV0 A0A026VY31 J9JNE6 A0A088A3U2 A0A158NRN0 A0A182R9R3 A0A182KUR6 A0A182X2X5 A0A084VMD9 A0A182UWL6 A0A182I4X9 A0A1S4H0U8 A0A1B6CN44 W5J8H7 A0A182YFF7 A0A195BLR4 A0A182TK73 A0A182NEU9 A0A2H8TM60 A0A182WGY1 A0A182SUA0 A0A232FB20 Q16S21 Q16S22 A0A1B6FZN6 A0A182LXN1 A0A023F2F5 T1HPP9 T1HLH4 E9RJV1 Q6DLV1 A7U8C8 A0A023ET86 A0A023ESV7 B4KKG6 A0A0T6AX31 A0A182JDS8 D6WLA0 B3MNP5 A0A0N0BH44 Q9NL84 A0A1S5QPR3 A0A067RL11 Q16GW0 Q16GW1 Q16895 A0A1Q3FVH7 A0A1J1HJ38 T1DQ67 A0A2A4IYP3 A0A1W4VXM7 J3JUQ3 A0A1D2NMP1 A0A2P8ZLL9 B4N186 A0A1A9V9K7 A0A1A9XGU4 W5J9T6 A0A1I8NE52 A0A1B6LPG7 A0A224XNZ0 A0A182F5H7 A0A0V0G4L0 A0A1L1ZLS9 A0A0Q5VXY7 M4GVV8 C3ZRR8 A0A2H1WW81 A0A1Y1KPJ8 H5V873 P18486 A0A0R1DPK1 A0A220K8N1 C9QP35 A0A3B5KIE6
EC Number
4.1.1.107
Pubmed
19121390
26354079
22651552
22118469
26227816
28004739
+ More
18362917 19820115 23537049 20075255 29403074 21719571 30249741 20798317 24508170 21347285 20966253 24438588 12364791 20920257 23761445 25244985 28648823 17510324 21283636 25474469 15926892 19366687 24945155 17994087 10785380 24845553 8673262 22516182 27289101 25315136 18563158 3021571 10731132 12537572 17550304 28630352 21551351
18362917 19820115 23537049 20075255 29403074 21719571 30249741 20798317 24508170 21347285 20966253 24438588 12364791 20920257 23761445 25244985 28648823 17510324 21283636 25474469 15926892 19366687 24945155 17994087 10785380 24845553 8673262 22516182 27289101 25315136 18563158 3021571 10731132 12537572 17550304 28630352 21551351
EMBL
BABH01010588
NWSH01000140
PCG79133.1
ODYU01005437
SOQ46283.1
KQ459601
+ More
KPI93764.1 AK401652 BAM18274.1 KQ461196 KPJ06526.1 AGBW02012749 OWR44502.1 JTDY01010491 KOB58186.1 GEZM01075053 JAV64312.1 GEZM01067891 JAV67338.1 GBYB01000239 GBYB01001421 JAG70006.1 JAG71188.1 KQ971343 EFA03414.1 NEVH01002553 PNF42051.1 APGK01040908 APGK01040909 APGK01040910 KB740984 ENN76340.1 PYGN01000022 PSN57405.1 KQ434787 KZC05332.1 GGMS01007563 MBY76766.1 AJVK01008775 AJVK01008776 AJVK01008777 AJWK01028413 KQ981931 KYN32806.1 GL888613 EGI59025.1 AXCN02000840 GGMR01001487 MBY14106.1 QOIP01000011 RLU17071.1 GL450531 EFN80849.1 KK107575 EZA48703.1 ABLF02017172 ADTU01024191 ATLV01014586 KE524975 KFB39133.1 APCN01003737 AAAB01008984 GEDC01022577 GEDC01017384 GEDC01014111 GEDC01012637 JAS14721.1 JAS19914.1 JAS23187.1 JAS24661.1 ADMH02001866 ETN60762.1 KQ976439 KYM86669.1 GFXV01003452 MBW15257.1 NNAY01000536 OXU27812.1 CH477689 EAT37247.1 EAT37246.1 GECZ01014133 JAS55636.1 AXCM01002775 GBBI01002915 JAC15797.1 ACPB03001467 ACPB03006673 AB618097 BAJ83478.1 AY662686 AAT75222.1 EU019710 ABU25222.1 GAPW01001063 JAC12535.1 GAPW01001220 JAC12378.1 CH933807 EDW11616.1 LJIG01022601 KRT79708.1 EFA03481.1 CH902620 EDV31132.1 KQ435759 KOX75704.1 AB028848 BAA95568.1 KU820948 AMQ13055.1 KK852451 KDR23678.1 CH478230 EAT33489.1 EAT33488.1 U27581 AAC31639.1 GFDL01003434 JAV31611.1 CVRI01000001 CRK86289.1 GAMD01001345 JAB00246.1 NWSH01005338 PCG64282.1 APGK01040898 APGK01040899 APGK01040900 APGK01040901 APGK01040902 APGK01040903 BT126968 AEE61930.1 ENN76335.1 LJIJ01000005 ODN06479.1 PSN57389.1 CH963920 EDW78026.2 ETN60761.1 GEBQ01014316 JAT25661.1 GFTR01006553 JAW09873.1 GECL01003504 JAP02620.1 KP250880 ALG64483.1 CH954179 KQS61906.1 JF795469 JF795470 AFG25780.1 AFG25781.1 GG666666 EEN44868.1 ODYU01011530 SOQ57325.1 GEZM01077299 JAV63333.1 BT133174 AEZ68802.1 X04695 AE014134 BT044421 CM000158 KRJ99144.1 MF319734 ASJ26448.1 BT099917 ACX32988.1
KPI93764.1 AK401652 BAM18274.1 KQ461196 KPJ06526.1 AGBW02012749 OWR44502.1 JTDY01010491 KOB58186.1 GEZM01075053 JAV64312.1 GEZM01067891 JAV67338.1 GBYB01000239 GBYB01001421 JAG70006.1 JAG71188.1 KQ971343 EFA03414.1 NEVH01002553 PNF42051.1 APGK01040908 APGK01040909 APGK01040910 KB740984 ENN76340.1 PYGN01000022 PSN57405.1 KQ434787 KZC05332.1 GGMS01007563 MBY76766.1 AJVK01008775 AJVK01008776 AJVK01008777 AJWK01028413 KQ981931 KYN32806.1 GL888613 EGI59025.1 AXCN02000840 GGMR01001487 MBY14106.1 QOIP01000011 RLU17071.1 GL450531 EFN80849.1 KK107575 EZA48703.1 ABLF02017172 ADTU01024191 ATLV01014586 KE524975 KFB39133.1 APCN01003737 AAAB01008984 GEDC01022577 GEDC01017384 GEDC01014111 GEDC01012637 JAS14721.1 JAS19914.1 JAS23187.1 JAS24661.1 ADMH02001866 ETN60762.1 KQ976439 KYM86669.1 GFXV01003452 MBW15257.1 NNAY01000536 OXU27812.1 CH477689 EAT37247.1 EAT37246.1 GECZ01014133 JAS55636.1 AXCM01002775 GBBI01002915 JAC15797.1 ACPB03001467 ACPB03006673 AB618097 BAJ83478.1 AY662686 AAT75222.1 EU019710 ABU25222.1 GAPW01001063 JAC12535.1 GAPW01001220 JAC12378.1 CH933807 EDW11616.1 LJIG01022601 KRT79708.1 EFA03481.1 CH902620 EDV31132.1 KQ435759 KOX75704.1 AB028848 BAA95568.1 KU820948 AMQ13055.1 KK852451 KDR23678.1 CH478230 EAT33489.1 EAT33488.1 U27581 AAC31639.1 GFDL01003434 JAV31611.1 CVRI01000001 CRK86289.1 GAMD01001345 JAB00246.1 NWSH01005338 PCG64282.1 APGK01040898 APGK01040899 APGK01040900 APGK01040901 APGK01040902 APGK01040903 BT126968 AEE61930.1 ENN76335.1 LJIJ01000005 ODN06479.1 PSN57389.1 CH963920 EDW78026.2 ETN60761.1 GEBQ01014316 JAT25661.1 GFTR01006553 JAW09873.1 GECL01003504 JAP02620.1 KP250880 ALG64483.1 CH954179 KQS61906.1 JF795469 JF795470 AFG25780.1 AFG25781.1 GG666666 EEN44868.1 ODYU01011530 SOQ57325.1 GEZM01077299 JAV63333.1 BT133174 AEZ68802.1 X04695 AE014134 BT044421 CM000158 KRJ99144.1 MF319734 ASJ26448.1 BT099917 ACX32988.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000007266 UP000235965 UP000019118 UP000002358 UP000245037 UP000076502 UP000092462 UP000092461 UP000078541 UP000007755 UP000075886 UP000279307 UP000008237 UP000053097 UP000007819 UP000005203 UP000005205 UP000075900 UP000075882 UP000076407 UP000030765 UP000075903 UP000075840 UP000000673 UP000076408 UP000078540 UP000075902 UP000075884 UP000075920 UP000075901 UP000215335 UP000008820 UP000075883 UP000015103 UP000009192 UP000075880 UP000007801 UP000053105 UP000027135 UP000183832 UP000192221 UP000094527 UP000007798 UP000078200 UP000092443 UP000095301 UP000069272 UP000008711 UP000001554 UP000000803 UP000002282 UP000005226
UP000007266 UP000235965 UP000019118 UP000002358 UP000245037 UP000076502 UP000092462 UP000092461 UP000078541 UP000007755 UP000075886 UP000279307 UP000008237 UP000053097 UP000007819 UP000005203 UP000005205 UP000075900 UP000075882 UP000076407 UP000030765 UP000075903 UP000075840 UP000000673 UP000076408 UP000078540 UP000075902 UP000075884 UP000075920 UP000075901 UP000215335 UP000008820 UP000075883 UP000015103 UP000009192 UP000075880 UP000007801 UP000053105 UP000027135 UP000183832 UP000192221 UP000094527 UP000007798 UP000078200 UP000092443 UP000095301 UP000069272 UP000008711 UP000001554 UP000000803 UP000002282 UP000005226
Pfam
Interpro
IPR015422
PyrdxlP-dep_Trfase_dom1
+ More
IPR010977 Aromatic_deC
IPR015424 PyrdxlP-dep_Trfase
IPR021115 Pyridoxal-P_BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR002129 PyrdxlP-dep_de-COase
IPR002937 Amino_oxidase
IPR036188 FAD/NAD-bd_sf
IPR011545 DEAD/DEAH_box_helicase_dom
IPR010240 Cys_deSase_IscS
IPR001650 Helicase_C
IPR020578 Aminotrans_V_PyrdxlP_BS
IPR000192 Aminotrans_V_dom
IPR014001 Helicase_ATP-bd
IPR007502 Helicase-assoc_dom
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR010977 Aromatic_deC
IPR015424 PyrdxlP-dep_Trfase
IPR021115 Pyridoxal-P_BS
IPR015421 PyrdxlP-dep_Trfase_major
IPR002129 PyrdxlP-dep_de-COase
IPR002937 Amino_oxidase
IPR036188 FAD/NAD-bd_sf
IPR011545 DEAD/DEAH_box_helicase_dom
IPR010240 Cys_deSase_IscS
IPR001650 Helicase_C
IPR020578 Aminotrans_V_PyrdxlP_BS
IPR000192 Aminotrans_V_dom
IPR014001 Helicase_ATP-bd
IPR007502 Helicase-assoc_dom
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
Gene 3D
CDD
ProteinModelPortal
H9J0C2
A0A2A4K5T6
A0A2H1VZS5
A0A194PM00
I4DK34
A0A194QSK8
+ More
A0A212ESM5 A0A0L7K538 A0A1Y1KSC4 A0A1Y1L3F9 A0A0C9QAN0 D6WLR5 A0A2J7RMJ6 N6U414 K7IPX7 A0A2P8ZLM8 A0A154P0I3 A0A2S2QGA6 A0A1B0DQW6 A0A1B0GK69 A0A195EXD1 F4X3J7 A0A182QMB0 A0A2S2NBF5 A0A3L8D9T2 E2BTV0 A0A026VY31 J9JNE6 A0A088A3U2 A0A158NRN0 A0A182R9R3 A0A182KUR6 A0A182X2X5 A0A084VMD9 A0A182UWL6 A0A182I4X9 A0A1S4H0U8 A0A1B6CN44 W5J8H7 A0A182YFF7 A0A195BLR4 A0A182TK73 A0A182NEU9 A0A2H8TM60 A0A182WGY1 A0A182SUA0 A0A232FB20 Q16S21 Q16S22 A0A1B6FZN6 A0A182LXN1 A0A023F2F5 T1HPP9 T1HLH4 E9RJV1 Q6DLV1 A7U8C8 A0A023ET86 A0A023ESV7 B4KKG6 A0A0T6AX31 A0A182JDS8 D6WLA0 B3MNP5 A0A0N0BH44 Q9NL84 A0A1S5QPR3 A0A067RL11 Q16GW0 Q16GW1 Q16895 A0A1Q3FVH7 A0A1J1HJ38 T1DQ67 A0A2A4IYP3 A0A1W4VXM7 J3JUQ3 A0A1D2NMP1 A0A2P8ZLL9 B4N186 A0A1A9V9K7 A0A1A9XGU4 W5J9T6 A0A1I8NE52 A0A1B6LPG7 A0A224XNZ0 A0A182F5H7 A0A0V0G4L0 A0A1L1ZLS9 A0A0Q5VXY7 M4GVV8 C3ZRR8 A0A2H1WW81 A0A1Y1KPJ8 H5V873 P18486 A0A0R1DPK1 A0A220K8N1 C9QP35 A0A3B5KIE6
A0A212ESM5 A0A0L7K538 A0A1Y1KSC4 A0A1Y1L3F9 A0A0C9QAN0 D6WLR5 A0A2J7RMJ6 N6U414 K7IPX7 A0A2P8ZLM8 A0A154P0I3 A0A2S2QGA6 A0A1B0DQW6 A0A1B0GK69 A0A195EXD1 F4X3J7 A0A182QMB0 A0A2S2NBF5 A0A3L8D9T2 E2BTV0 A0A026VY31 J9JNE6 A0A088A3U2 A0A158NRN0 A0A182R9R3 A0A182KUR6 A0A182X2X5 A0A084VMD9 A0A182UWL6 A0A182I4X9 A0A1S4H0U8 A0A1B6CN44 W5J8H7 A0A182YFF7 A0A195BLR4 A0A182TK73 A0A182NEU9 A0A2H8TM60 A0A182WGY1 A0A182SUA0 A0A232FB20 Q16S21 Q16S22 A0A1B6FZN6 A0A182LXN1 A0A023F2F5 T1HPP9 T1HLH4 E9RJV1 Q6DLV1 A7U8C8 A0A023ET86 A0A023ESV7 B4KKG6 A0A0T6AX31 A0A182JDS8 D6WLA0 B3MNP5 A0A0N0BH44 Q9NL84 A0A1S5QPR3 A0A067RL11 Q16GW0 Q16GW1 Q16895 A0A1Q3FVH7 A0A1J1HJ38 T1DQ67 A0A2A4IYP3 A0A1W4VXM7 J3JUQ3 A0A1D2NMP1 A0A2P8ZLL9 B4N186 A0A1A9V9K7 A0A1A9XGU4 W5J9T6 A0A1I8NE52 A0A1B6LPG7 A0A224XNZ0 A0A182F5H7 A0A0V0G4L0 A0A1L1ZLS9 A0A0Q5VXY7 M4GVV8 C3ZRR8 A0A2H1WW81 A0A1Y1KPJ8 H5V873 P18486 A0A0R1DPK1 A0A220K8N1 C9QP35 A0A3B5KIE6
PDB
6JRL
E-value=3.93775e-150,
Score=1364
Ontologies
PATHWAY
GO
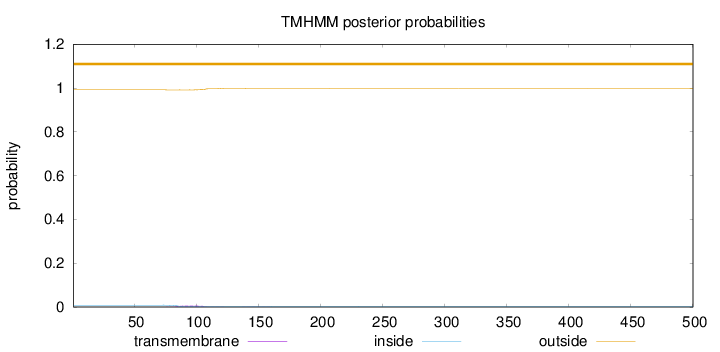
Topology
Length:
500
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.161460000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00836
outside
1 - 500
Population Genetic Test Statistics
Pi
5.929071
Theta
18.388734
Tajima's D
-1.353421
CLR
1.102607
CSRT
0.0785460726963652
Interpretation
Uncertain
