Gene
KWMTBOMO02028
Pre Gene Modal
BGIBMGA002956
Annotation
PREDICTED:_protein_phosphatase_PHLPP-like_protein_isoform_X2_[Bombyx_mori]
Full name
Protein phosphatase PHLPP-like protein
Alternative Name
PH domain leucine-rich repeat protein phosphatase
dPHLPP
dPHLPP
Location in the cell
Nuclear Reliability : 2.97
Sequence
CDS
ATGGCGCCGGATGAAGAAAATAAAGTTGAATATCGTACTCCACTAACCAATGTTGATTTACGTCACAATAGATTAAAAGGAAGCATAATACTCGGAAATTATGAGCATTTAGTATGTTTAGATATATCGCATAATTCTGTGGAAGTTTTAGTCTTGTCTTCACTGCGGGGACTTCGGGAACTATACGCTTCACATAATGCACTCCAGCACGTGGCGCTTCATGGAGGCTCTTTACGAATTCTCCGAGCCCCCCACAATCAATTAGAATCATTGACAAGTACAGTCCCACCAATCAATTTAGTAGAAATCGACGTGTCATACAACAAATTGACGTTACTACCACCATGGATCAGCGGCTGTTCCGACCTCACCACGCTTCACGCGAGCAATAACCAATTAACCTCACTTCCAGACCATTTGTTCTGTAGTGAACTTTCAAGTCTGAGTAACCTGCATTTAGCCCATAATAAAATCTCTAGTATACCATCAATGCCGCGCCTAAGAGCTCCCCTTAAAGAATTACTGCTTCATGATAACTCTATTCAGTCACTGCCAGAAAACTTCTTTTCGGTATGTGACAGATTGGGCGTTTTAAATCTTTCGAATAACAGACTGAGCCGCTTACCTTTGGCAAGATGTACCTCATATTGTCTAGAGTATCTATACCTAACAGCTAACTGCTTGAATGACGATGCAACAGATGTTCTCATAGCATTCCGTGGCTTAAAGATTCTACATTTGGCCTACAACTGTCTAACGCGGCTTCCAGAAAACTGTGTCAACTATTGGCCCGATGTTGAAGAACTAGTACTCTCGGGGAACCTGTTTTCAAGATTGCCAGAAAATATGCCTCAGCTCAATAACATCAGAGTCGTAAGGGCTCACTCAAATAAACTACGTTCGGTACCTATGTTCGCATGTAGCGCTTCGGTGAAGATTTTAGATTTTGCTCACAACGAACTGGATAGGGTAGATTTACGGCTACTTGCACCAAAACAATTAAAATTTCTAGATATATCTTGTAACAAAAAATTACAAATGGACCCTTCACAGTTTAATTCCTATAAATGTCAAAGACCATTGAGCTTAGTAGATGTGACAGGGCGACGTTCAGTTTCGTGGACCCACAAAACAGGATATCACGAAGAATTAACTGGCATGACCCCGTGGAATACAGGATTTTCTGAATGCCCGGACAAATCATTGCAGTTATATTGTGCACAAATACGTCTTCCTTCGTTTTGTAATAAAGAGGGCCTTTTCGCCGTCATTGATGGTGAAACTGATATCGAAGTACCAAAAATTTTACAAACCAGTATACCAGGATTAGTTCTTGAAGAGAAATCAATCAAAGAAACTGTTAACGAATATATGAAATACGTAGTATTATCGGCTCATAGAGAATTGAAAGAAAAAGGTCAAAGAAAAGGAGCCTGCATGGTCATGTGCCACTTGTCTCCTATTAATGTACAAGAAAATGCATTCGGACAGGCGTTAAAAAAATATAGTTTAAAATTAGCTAACGTAGGCGATACTAGAGCAGTTTTAAGTAGACGTAATGGACCGTTGAGTTTGGGCATAGAAAATAAAAGACGTCTAGGATATTCCTGTCGCTATCCCAATACTGTCCCAGATCCGGATATCATCCAGACAGTTATTAAAGAAGACGACGAATTTCTGATATTAGCCAATGGGAAATTCTGGGGTGCTGTTAGTATAGATTCTGCAGTTTCTGAAGTAAGAGCAGAACGAAATCCTGTTCTTGCAGCAAAAAGATTACAAGACCTGGCCCAAAGCTATGGCATCGAAGAATGTATTTCAATTATAGTAATTAGGTTTGATAATGAAAGAGCTGATGTTGACCTTTTGATGAGAGAATTAAGGCAGACGATTAACGGGAATAAAACGACATGTCGATCGGATTGTTGTTGTTCACGTTTGGAACCTTGCTGTCACTCAATATCTCCGTCAAAGCCGAGCAGCGATCGATCTTCGCCAAGCGGGCAAAGTGACCGCCCTCTAAGTGAAGCAATAAACCATCAACATTACTCCAGTGTTCGATCGCAAAATAAAGCATCCGAAAGACGGAGCTGTCGCGGAGGCGTGGCTAGAGCAATTCGTGTTCGTGTTGAAGAAGAGAAAGAAGATCACCGAAATGAGGAAACTTCAAATCCAGAGGAACAATTTAAATGCTGGGAATACATGTTGGAACAGAACACTCAAATGCTGTTCGACAAGGAATTAGATAACTTATCGAAGGGTATTCGTTCGAATGGAAGTACTCTGAGGAACACAAAAGGGTTGTCGGGGAGTAGCCCTCAATTACATTTAAGTGGAAAAACTTCGAAAGTTCCATTCTTGTCTAAACATTTTGGTAGTGCAAGATCTTTTGGCACAGCTTTAAAGCCTGACTTTCGATTCGGCTCTGGAAGATTGCCGAATGGTGGTCCTAACGCTGCATATTTTGGTTCACTTCAACGACTCATGCCTTATCATTTAGAATATGACTTTGCCGTAATTCAAGAGAAAACAACACACTCCCAAGACTCGCTCGACCTCGAAGGAAGAATGTCACAATATTGGGGAGTCGCTACAACCGAATTATAA
Protein
MAPDEENKVEYRTPLTNVDLRHNRLKGSIILGNYEHLVCLDISHNSVEVLVLSSLRGLRELYASHNALQHVALHGGSLRILRAPHNQLESLTSTVPPINLVEIDVSYNKLTLLPPWISGCSDLTTLHASNNQLTSLPDHLFCSELSSLSNLHLAHNKISSIPSMPRLRAPLKELLLHDNSIQSLPENFFSVCDRLGVLNLSNNRLSRLPLARCTSYCLEYLYLTANCLNDDATDVLIAFRGLKILHLAYNCLTRLPENCVNYWPDVEELVLSGNLFSRLPENMPQLNNIRVVRAHSNKLRSVPMFACSASVKILDFAHNELDRVDLRLLAPKQLKFLDISCNKKLQMDPSQFNSYKCQRPLSLVDVTGRRSVSWTHKTGYHEELTGMTPWNTGFSECPDKSLQLYCAQIRLPSFCNKEGLFAVIDGETDIEVPKILQTSIPGLVLEEKSIKETVNEYMKYVVLSAHRELKEKGQRKGACMVMCHLSPINVQENAFGQALKKYSLKLANVGDTRAVLSRRNGPLSLGIENKRRLGYSCRYPNTVPDPDIIQTVIKEDDEFLILANGKFWGAVSIDSAVSEVRAERNPVLAAKRLQDLAQSYGIEECISIIVIRFDNERADVDLLMRELRQTINGNKTTCRSDCCCSRLEPCCHSISPSKPSSDRSSPSGQSDRPLSEAINHQHYSSVRSQNKASERRSCRGGVARAIRVRVEEEKEDHRNEETSNPEEQFKCWEYMLEQNTQMLFDKELDNLSKGIRSNGSTLRNTKGLSGSSPQLHLSGKTSKVPFLSKHFGSARSFGTALKPDFRFGSGRLPNGGPNAAYFGSLQRLMPYHLEYDFAVIQEKTTHSQDSLDLEGRMSQYWGVATTEL
Summary
Description
Protein phosphatase that specifically mediates dephosphorylation of 'Ser-586' of Akt1, a protein that regulates the balance between cell survival and apoptosis through a cascade that primarily alters the function of transcription factors that regulate pro- and antiapoptotic genes. Dephosphorylation of 'Ser-586' of Akt1 triggers apoptosis and suppression of tumor growth.
Catalytic Activity
H2O + O-phospho-L-seryl-[protein] = L-seryl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
H2O + O-phospho-L-threonyl-[protein] = L-threonyl-[protein] + phosphate
Cofactor
Mn(2+)
Similarity
Belongs to the CRISP family.
Keywords
Apoptosis
Complete proteome
Hydrolase
Leucine-rich repeat
Manganese
Metal-binding
Protein phosphatase
Reference proteome
Repeat
Feature
chain Protein phosphatase PHLPP-like protein
Uniprot
H9J0C0
A0A2H1W8Z9
A0A2A4JXX1
A0A0L7LKN1
A0A194R8K4
A0A194PK03
+ More
A0A212ESM7 D6WLS2 A0A1B0DQW8 A0A1Y1LIB7 A0A1S4H022 A0A182I4X6 A0A182U465 A0A182KUS7 A0A182X2X8 A0A182UZM4 Q7PW55 A0A182K9A3 A0A182SHF0 A0A182QHB5 A0A182YFG4 A0A182IWW6 A0A0A9XBC9 A0A182NEV3 A0A084VMD4 N6TAC1 A0A2J7RAW6 U5EXY9 A0A336M0J4 A0A336KQA3 A0A067R0H2 A0A2P8ZIF3 B0X2D7 A0A182M9I8 W5JCZ5 A0A1J1HZP3 E2ACQ4 F4W688 E2BUY5 A0A151WHU8 A0A182F5I1 A0A3L8DDB4 A0A195FJ86 A0A182R9R6 A0A151ILH3 A0A182WGX8 A0A195DMH4 A0A182PDE7 Q173L1 A0A0N0BED4 A0A088ACL7 A0A310SPR8 T1GVH3 A0A026WA02 A0A158P2K6 A0A195AW70 A0A1I8P3Y6 A0A0A9XEP9 T1HYD9 A0A0L7QZ03 A0A1B0GK67 A0A154PJC9 B4LS13 B4KE63 A0A0M4E297 B3MNQ6 A0A0J9R5D9 B4Q9F6 A0A0L0BXF2 B4I5Q6 M9PBD5 Q9VJ07 B3NMA1 B4PAG1 B4N146 A0A3Q0J5Q0 A0A1W4VXJ6 A0A3B0K0N3 K7J905 A0A0K8WI19 A0A1B0F9U0 A0A1A9XGV0 A0A1B0BJG8 A0A1A9W036 B4JAM4 A0A1A9V9L5 A0A1B0A513 A0A1I8NFV6 A0A2A3ENE2 Q29PI5 B4GKT8 K1QUE9
A0A212ESM7 D6WLS2 A0A1B0DQW8 A0A1Y1LIB7 A0A1S4H022 A0A182I4X6 A0A182U465 A0A182KUS7 A0A182X2X8 A0A182UZM4 Q7PW55 A0A182K9A3 A0A182SHF0 A0A182QHB5 A0A182YFG4 A0A182IWW6 A0A0A9XBC9 A0A182NEV3 A0A084VMD4 N6TAC1 A0A2J7RAW6 U5EXY9 A0A336M0J4 A0A336KQA3 A0A067R0H2 A0A2P8ZIF3 B0X2D7 A0A182M9I8 W5JCZ5 A0A1J1HZP3 E2ACQ4 F4W688 E2BUY5 A0A151WHU8 A0A182F5I1 A0A3L8DDB4 A0A195FJ86 A0A182R9R6 A0A151ILH3 A0A182WGX8 A0A195DMH4 A0A182PDE7 Q173L1 A0A0N0BED4 A0A088ACL7 A0A310SPR8 T1GVH3 A0A026WA02 A0A158P2K6 A0A195AW70 A0A1I8P3Y6 A0A0A9XEP9 T1HYD9 A0A0L7QZ03 A0A1B0GK67 A0A154PJC9 B4LS13 B4KE63 A0A0M4E297 B3MNQ6 A0A0J9R5D9 B4Q9F6 A0A0L0BXF2 B4I5Q6 M9PBD5 Q9VJ07 B3NMA1 B4PAG1 B4N146 A0A3Q0J5Q0 A0A1W4VXJ6 A0A3B0K0N3 K7J905 A0A0K8WI19 A0A1B0F9U0 A0A1A9XGV0 A0A1B0BJG8 A0A1A9W036 B4JAM4 A0A1A9V9L5 A0A1B0A513 A0A1I8NFV6 A0A2A3ENE2 Q29PI5 B4GKT8 K1QUE9
EC Number
3.1.3.16
Pubmed
19121390
26227816
26354079
22118469
18362917
19820115
+ More
28004739 12364791 20966253 25244985 25401762 24438588 23537049 24845553 29403074 20920257 23761445 20798317 21719571 30249741 17510324 24508170 21347285 17994087 18057021 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15808505 17550304 20075255 25315136 15632085 23185243 22992520
28004739 12364791 20966253 25244985 25401762 24438588 23537049 24845553 29403074 20920257 23761445 20798317 21719571 30249741 17510324 24508170 21347285 17994087 18057021 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 15808505 17550304 20075255 25315136 15632085 23185243 22992520
EMBL
BABH01010595
BABH01010596
ODYU01007084
SOQ49540.1
NWSH01000407
PCG76616.1
+ More
JTDY01000830 KOB75746.1 KQ460597 KPJ13834.1 KQ459601 KPI93766.1 AGBW02012749 OWR44506.1 KQ971343 EFA04777.2 AJVK01008787 GEZM01054811 JAV73384.1 AAAB01008984 APCN01003737 EAA14746.4 AXCN02000840 GBHO01026305 JAG17299.1 ATLV01014585 ATLV01014586 KE524975 KFB39128.1 APGK01037103 APGK01037104 APGK01037105 KB740948 ENN77219.1 NEVH01006564 PNF37972.1 GANO01000670 JAB59201.1 UFQT01000222 SSX21947.1 UFQS01000634 UFQT01000634 SSX05605.1 SSX25964.1 KK852804 KDR16236.1 PYGN01000048 PSN56260.1 DS232286 EDS39160.1 AXCM01002871 ADMH02001866 ETN60765.1 CVRI01000037 CRK93493.1 GL438568 EFN68798.1 GL887707 EGI70223.1 GL450772 EFN80522.1 KQ983112 KYQ47375.1 QOIP01000010 RLU18152.1 KQ981523 KYN40431.1 KQ977120 KYN05594.1 KQ980724 KYN14100.1 CH477422 EAT41218.2 KQ435840 KOX71414.1 KQ761289 OAD57914.1 CAQQ02194901 KK107310 EZA52875.1 ADTU01006944 ADTU01006945 ADTU01006946 ADTU01006947 ADTU01006948 ADTU01006949 ADTU01006950 ADTU01006951 KQ976731 KYM76280.1 GBHO01026306 JAG17298.1 ACPB03007941 KQ414685 KOC63792.1 AJWK01028404 AJWK01028405 KQ434936 KZC11955.1 CH940649 EDW63689.2 KRF81248.1 CH933807 EDW11808.1 CP012523 ALC39678.1 CH902620 EDV31143.1 CM002910 KMY90934.1 CM000361 EDX05463.1 JRES01001289 KNC23914.1 CH480822 EDW55712.1 AE014134 AGB93093.1 CH954179 EDV54701.1 CM000158 EDW90369.1 CH963920 EDW78136.1 OUUW01000010 SPP86222.1 GDHF01001518 JAI50796.1 CCAG010003100 JXJN01015387 CH916368 EDW02810.1 KZ288204 PBC33237.1 CH379058 EAL34308.2 KRT03375.1 CH479184 EDW37254.1 JH816884 EKC25171.1
JTDY01000830 KOB75746.1 KQ460597 KPJ13834.1 KQ459601 KPI93766.1 AGBW02012749 OWR44506.1 KQ971343 EFA04777.2 AJVK01008787 GEZM01054811 JAV73384.1 AAAB01008984 APCN01003737 EAA14746.4 AXCN02000840 GBHO01026305 JAG17299.1 ATLV01014585 ATLV01014586 KE524975 KFB39128.1 APGK01037103 APGK01037104 APGK01037105 KB740948 ENN77219.1 NEVH01006564 PNF37972.1 GANO01000670 JAB59201.1 UFQT01000222 SSX21947.1 UFQS01000634 UFQT01000634 SSX05605.1 SSX25964.1 KK852804 KDR16236.1 PYGN01000048 PSN56260.1 DS232286 EDS39160.1 AXCM01002871 ADMH02001866 ETN60765.1 CVRI01000037 CRK93493.1 GL438568 EFN68798.1 GL887707 EGI70223.1 GL450772 EFN80522.1 KQ983112 KYQ47375.1 QOIP01000010 RLU18152.1 KQ981523 KYN40431.1 KQ977120 KYN05594.1 KQ980724 KYN14100.1 CH477422 EAT41218.2 KQ435840 KOX71414.1 KQ761289 OAD57914.1 CAQQ02194901 KK107310 EZA52875.1 ADTU01006944 ADTU01006945 ADTU01006946 ADTU01006947 ADTU01006948 ADTU01006949 ADTU01006950 ADTU01006951 KQ976731 KYM76280.1 GBHO01026306 JAG17298.1 ACPB03007941 KQ414685 KOC63792.1 AJWK01028404 AJWK01028405 KQ434936 KZC11955.1 CH940649 EDW63689.2 KRF81248.1 CH933807 EDW11808.1 CP012523 ALC39678.1 CH902620 EDV31143.1 CM002910 KMY90934.1 CM000361 EDX05463.1 JRES01001289 KNC23914.1 CH480822 EDW55712.1 AE014134 AGB93093.1 CH954179 EDV54701.1 CM000158 EDW90369.1 CH963920 EDW78136.1 OUUW01000010 SPP86222.1 GDHF01001518 JAI50796.1 CCAG010003100 JXJN01015387 CH916368 EDW02810.1 KZ288204 PBC33237.1 CH379058 EAL34308.2 KRT03375.1 CH479184 EDW37254.1 JH816884 EKC25171.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000092462 UP000075840 UP000075902 UP000075882 UP000076407 UP000075903 UP000007062 UP000075881 UP000075901 UP000075886 UP000076408 UP000075880 UP000075884 UP000030765 UP000019118 UP000235965 UP000027135 UP000245037 UP000002320 UP000075883 UP000000673 UP000183832 UP000000311 UP000007755 UP000008237 UP000075809 UP000069272 UP000279307 UP000078541 UP000075900 UP000078542 UP000075920 UP000078492 UP000075885 UP000008820 UP000053105 UP000005203 UP000015102 UP000053097 UP000005205 UP000078540 UP000095300 UP000015103 UP000053825 UP000092461 UP000076502 UP000008792 UP000009192 UP000092553 UP000007801 UP000000304 UP000037069 UP000001292 UP000000803 UP000008711 UP000002282 UP000007798 UP000079169 UP000192221 UP000268350 UP000002358 UP000092444 UP000092443 UP000092460 UP000091820 UP000001070 UP000078200 UP000092445 UP000095301 UP000242457 UP000001819 UP000008744 UP000005408
UP000007266 UP000092462 UP000075840 UP000075902 UP000075882 UP000076407 UP000075903 UP000007062 UP000075881 UP000075901 UP000075886 UP000076408 UP000075880 UP000075884 UP000030765 UP000019118 UP000235965 UP000027135 UP000245037 UP000002320 UP000075883 UP000000673 UP000183832 UP000000311 UP000007755 UP000008237 UP000075809 UP000069272 UP000279307 UP000078541 UP000075900 UP000078542 UP000075920 UP000078492 UP000075885 UP000008820 UP000053105 UP000005203 UP000015102 UP000053097 UP000005205 UP000078540 UP000095300 UP000015103 UP000053825 UP000092461 UP000076502 UP000008792 UP000009192 UP000092553 UP000007801 UP000000304 UP000037069 UP000001292 UP000000803 UP000008711 UP000002282 UP000007798 UP000079169 UP000192221 UP000268350 UP000002358 UP000092444 UP000092443 UP000092460 UP000091820 UP000001070 UP000078200 UP000092445 UP000095301 UP000242457 UP000001819 UP000008744 UP000005408
Pfam
Interpro
IPR032675
LRR_dom_sf
+ More
IPR036457 PPM-type_dom_sf
IPR001932 PPM-type_phosphatase_dom
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR014044 CAP_domain
IPR013871 Cysteine_rich_secretory
IPR035940 CAP_sf
IPR001283 CRISP-related
IPR002413 V5_allergen-like
IPR018244 Allrgn_V5/Tpx1_CS
IPR040676 DUF5641
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR011993 PH-like_dom_sf
IPR001849 PH_domain
IPR036457 PPM-type_dom_sf
IPR001932 PPM-type_phosphatase_dom
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR014044 CAP_domain
IPR013871 Cysteine_rich_secretory
IPR035940 CAP_sf
IPR001283 CRISP-related
IPR002413 V5_allergen-like
IPR018244 Allrgn_V5/Tpx1_CS
IPR040676 DUF5641
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR019786 Zinc_finger_PHD-type_CS
IPR011993 PH-like_dom_sf
IPR001849 PH_domain
CDD
ProteinModelPortal
H9J0C0
A0A2H1W8Z9
A0A2A4JXX1
A0A0L7LKN1
A0A194R8K4
A0A194PK03
+ More
A0A212ESM7 D6WLS2 A0A1B0DQW8 A0A1Y1LIB7 A0A1S4H022 A0A182I4X6 A0A182U465 A0A182KUS7 A0A182X2X8 A0A182UZM4 Q7PW55 A0A182K9A3 A0A182SHF0 A0A182QHB5 A0A182YFG4 A0A182IWW6 A0A0A9XBC9 A0A182NEV3 A0A084VMD4 N6TAC1 A0A2J7RAW6 U5EXY9 A0A336M0J4 A0A336KQA3 A0A067R0H2 A0A2P8ZIF3 B0X2D7 A0A182M9I8 W5JCZ5 A0A1J1HZP3 E2ACQ4 F4W688 E2BUY5 A0A151WHU8 A0A182F5I1 A0A3L8DDB4 A0A195FJ86 A0A182R9R6 A0A151ILH3 A0A182WGX8 A0A195DMH4 A0A182PDE7 Q173L1 A0A0N0BED4 A0A088ACL7 A0A310SPR8 T1GVH3 A0A026WA02 A0A158P2K6 A0A195AW70 A0A1I8P3Y6 A0A0A9XEP9 T1HYD9 A0A0L7QZ03 A0A1B0GK67 A0A154PJC9 B4LS13 B4KE63 A0A0M4E297 B3MNQ6 A0A0J9R5D9 B4Q9F6 A0A0L0BXF2 B4I5Q6 M9PBD5 Q9VJ07 B3NMA1 B4PAG1 B4N146 A0A3Q0J5Q0 A0A1W4VXJ6 A0A3B0K0N3 K7J905 A0A0K8WI19 A0A1B0F9U0 A0A1A9XGV0 A0A1B0BJG8 A0A1A9W036 B4JAM4 A0A1A9V9L5 A0A1B0A513 A0A1I8NFV6 A0A2A3ENE2 Q29PI5 B4GKT8 K1QUE9
A0A212ESM7 D6WLS2 A0A1B0DQW8 A0A1Y1LIB7 A0A1S4H022 A0A182I4X6 A0A182U465 A0A182KUS7 A0A182X2X8 A0A182UZM4 Q7PW55 A0A182K9A3 A0A182SHF0 A0A182QHB5 A0A182YFG4 A0A182IWW6 A0A0A9XBC9 A0A182NEV3 A0A084VMD4 N6TAC1 A0A2J7RAW6 U5EXY9 A0A336M0J4 A0A336KQA3 A0A067R0H2 A0A2P8ZIF3 B0X2D7 A0A182M9I8 W5JCZ5 A0A1J1HZP3 E2ACQ4 F4W688 E2BUY5 A0A151WHU8 A0A182F5I1 A0A3L8DDB4 A0A195FJ86 A0A182R9R6 A0A151ILH3 A0A182WGX8 A0A195DMH4 A0A182PDE7 Q173L1 A0A0N0BED4 A0A088ACL7 A0A310SPR8 T1GVH3 A0A026WA02 A0A158P2K6 A0A195AW70 A0A1I8P3Y6 A0A0A9XEP9 T1HYD9 A0A0L7QZ03 A0A1B0GK67 A0A154PJC9 B4LS13 B4KE63 A0A0M4E297 B3MNQ6 A0A0J9R5D9 B4Q9F6 A0A0L0BXF2 B4I5Q6 M9PBD5 Q9VJ07 B3NMA1 B4PAG1 B4N146 A0A3Q0J5Q0 A0A1W4VXJ6 A0A3B0K0N3 K7J905 A0A0K8WI19 A0A1B0F9U0 A0A1A9XGV0 A0A1B0BJG8 A0A1A9W036 B4JAM4 A0A1A9V9L5 A0A1B0A513 A0A1I8NFV6 A0A2A3ENE2 Q29PI5 B4GKT8 K1QUE9
PDB
5A5C
E-value=1.60712e-11,
Score=171
Ontologies
GO
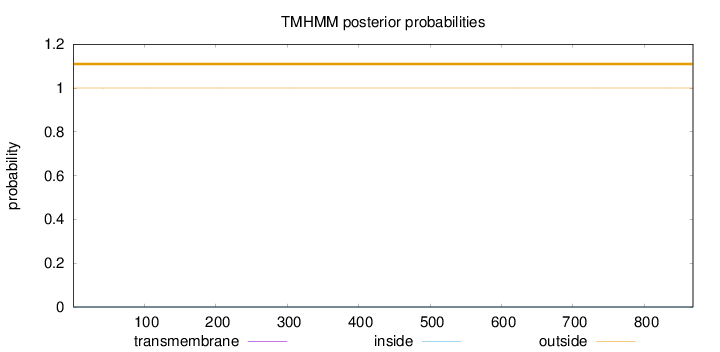
Topology
Length:
868
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00301
Exp number, first 60 AAs:
0.00221
Total prob of N-in:
0.00016
outside
1 - 868
Population Genetic Test Statistics
Pi
70.154016
Theta
176.57484
Tajima's D
-2.016088
CLR
0.516297
CSRT
0.0140492975351232
Interpretation
Uncertain
