Gene
KWMTBOMO02027
Pre Gene Modal
BGIBMGA002955
Annotation
PREDICTED:_protein_phosphatase_PHLPP-like_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.33
Sequence
CDS
ATGCTTCTCCGATGTGTAGTATGGTTACAATTTATACTGATTCTAGATTTGAGTGGACATTTGCATACTGAGAAAGTTTTGGATAAGGATAAACAATTATTTCCACCTCATCTCATCGGAGAAGATTCGCTGAACCCTAAAAGGGAAATTGTTCGGAGAAAAATAGTACTTTATCATAATTTTTTCCGTTCCAAAGTTCGACCAATTGCGTCGAATATGCTCCTCATGTCGTGGCATCCCAAGGCAGCTCGTCAAGCACAGCGCTACGCAGAGCAATGCGTGTATTTAACACATAATGATATTAAAGAGAACACCGTACCTTCTCTGGGAACCTGTGGACAAAATTTATTCGTAGCGGCACAGAAGACACCATGGTTTTTCGCCATAAAGACTTGGTTCGTGGAGAATGAAAACTTCACGTATGGAGATCCAAACGTGAACCTCACAGCAGTAGGACACTACACCCAAATGGTTTGGGCAACGAGCTACAAAGTCGGATGTGGTCTAGCGCATTGTTCCGGGGGACCATGGGGACATTTTTATAACTACGTGTGTCACTATTGTCCTGGTGGCAACATCCTCACAATAGCACAGTATCCATACAAAACTGGTGAGAAATGCTCGGATTGCCCAGATCACTGCACATCTGAGTCTTTGTGCACGAATTTCTGTCCGGTTCGTGACTATTACTCTAACTGCGATGAGCTAGTAAAAAATATCGACGGTTTCTGCAAGCAAGGAATGTGCAACGCGACTTGTTCATGTGGATCGAAACTTCTTCACAAGAATTATCCTTGCAGCGGCGGGCGCAGAGTCCGGCGTGCGGCCATTGCGTCGTCGGACATCGTACCTCGCGAGCACCTGCCTCCGCGCGCCATGGTGTGTGATGATAGCTTGCATGCTTCATCGCCTCACGTGCGTCGCAAGTCACTTCGCCGTCTTGCATCTACTCGCGGCTTTAAGTCTCGTAATCAAGACGAACACGGCTGGATCAGAGTTTTCGAAGGCCTTGAACCCTTCGCAGTTGATGCACCCAGTAAATTGGTGAAAGTTACTTTGAGTACAACTGTTGAAGAAATCAACAAAGACCTTGGGTTTACTGATGAATCAACGTTATGGATGCAAATTGGTGGGGAAAAATCGAGACGGCTTGAACTTGATGAATACCCCCTAAGGATACAAGACCAATTTTTAGCAACATGTGGATGGAGATCAGAATCTCGAAGAAAACGTCTAGCTGTTGATCCTGAGTTGCGACAGTCATTACGATGGTGTGCTGGACCAGCCGGAAATCATGGAGGTGTTTTGAGATCTGGAACTTTATATGTTTTAAAAGGACACGTTTTCCCACAATGGAAACCGAAACCTGCATTTATCATTGGGTCAAGGCTGCACACGTATGGAACTACTTGGGAGATGCTGGAGTTAAGCGGAGGAAACATAGACTTCTGCCCCCCAAAAGCGCAAAGACTTGTGTTGTGCGTAAAGCCCCGGCCAAGTAATAACTTGCCATGTGACAATAATGCAAATCACGTCTTCTTAGGATTCAGCACCGTTTGGGAAAGGAACATTTGGTTCAGCTGGCTCAAAGAACTCTAA
Protein
MLLRCVVWLQFILILDLSGHLHTEKVLDKDKQLFPPHLIGEDSLNPKREIVRRKIVLYHNFFRSKVRPIASNMLLMSWHPKAARQAQRYAEQCVYLTHNDIKENTVPSLGTCGQNLFVAAQKTPWFFAIKTWFVENENFTYGDPNVNLTAVGHYTQMVWATSYKVGCGLAHCSGGPWGHFYNYVCHYCPGGNILTIAQYPYKTGEKCSDCPDHCTSESLCTNFCPVRDYYSNCDELVKNIDGFCKQGMCNATCSCGSKLLHKNYPCSGGRRVRRAAIASSDIVPREHLPPRAMVCDDSLHASSPHVRRKSLRRLASTRGFKSRNQDEHGWIRVFEGLEPFAVDAPSKLVKVTLSTTVEEINKDLGFTDESTLWMQIGGEKSRRLELDEYPLRIQDQFLATCGWRSESRRKRLAVDPELRQSLRWCAGPAGNHGGVLRSGTLYVLKGHVFPQWKPKPAFIIGSRLHTYGTTWEMLELSGGNIDFCPPKAQRLVLCVKPRPSNNLPCDNNANHVFLGFSTVWERNIWFSWLKEL
Summary
Similarity
Belongs to the CRISP family.
Uniprot
EMBL
Proteomes
Interpro
IPR036457
PPM-type_dom_sf
+ More
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR014044 CAP_domain
IPR003591 Leu-rich_rpt_typical-subtyp
IPR013871 Cysteine_rich_secretory
IPR035940 CAP_sf
IPR001283 CRISP-related
IPR002413 V5_allergen-like
IPR001932 PPM-type_phosphatase_dom
IPR018244 Allrgn_V5/Tpx1_CS
IPR032675 LRR_dom_sf
IPR001611 Leu-rich_rpt
IPR014044 CAP_domain
IPR003591 Leu-rich_rpt_typical-subtyp
IPR013871 Cysteine_rich_secretory
IPR035940 CAP_sf
IPR001283 CRISP-related
IPR002413 V5_allergen-like
IPR001932 PPM-type_phosphatase_dom
IPR018244 Allrgn_V5/Tpx1_CS
Gene 3D
CDD
ProteinModelPortal
PDB
6IMF
E-value=8.49651e-40,
Score=412
Ontologies
KEGG
GO
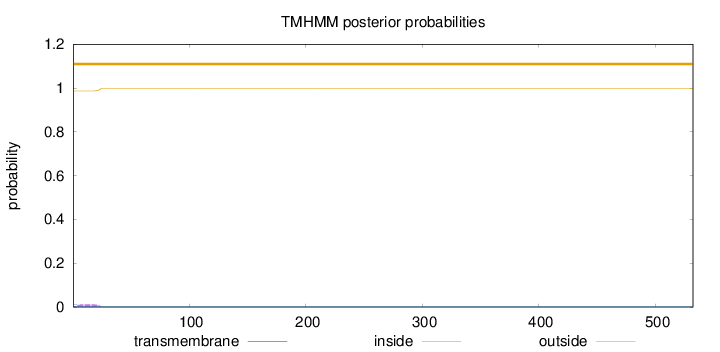
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.835486

Length:
532
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.234630000000001
Exp number, first 60 AAs:
0.23016
Total prob of N-in:
0.01299
outside
1 - 532
Population Genetic Test Statistics
Pi
268.184288
Theta
210.148282
Tajima's D
0.756354
CLR
0.084657
CSRT
0.591470426478676
Interpretation
Uncertain
